Difference between revisions of "GotCloud"
| Line 47: | Line 47: | ||
After completing the GotCloud Variant Calling PIpeline, [[EPACTS|EPACTS (Efficient and Parallelizable Association Container Toolbox)]] can be used to perform statistical tests to identify genome-wide association from sequence data. | After completing the GotCloud Variant Calling PIpeline, [[EPACTS|EPACTS (Efficient and Parallelizable Association Container Toolbox)]] can be used to perform statistical tests to identify genome-wide association from sequence data. | ||
| − | [[File:GotCloudDiagram. | + | [[File:GotCloudDiagram.jpg]] |
Revision as of 14:34, 18 March 2013
Genomes on the Cloud (GotCloud) Introduction
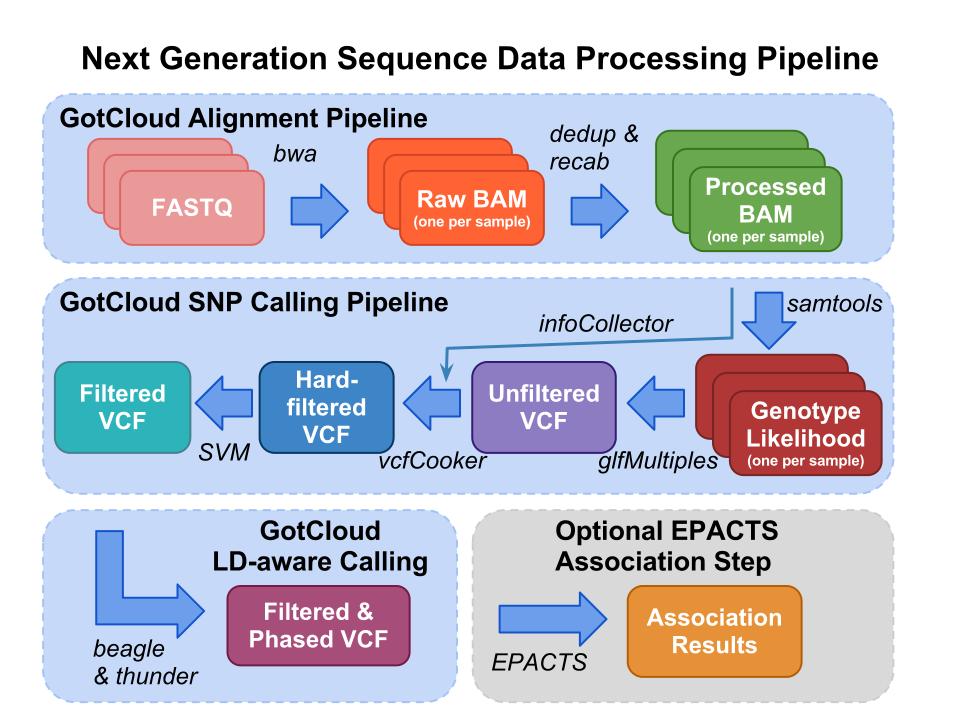
To handle the increasing volume of next generation sequencing and genotyping data, we created and developed software pipelines called Genomes on the Cloud (GotCloud).
GotCloud contains Mapping & Variant Calling Pipelines.
Key Features:
- Connects sequence analysis tools together in automated pipeline
- Alignment, quality control, variant calling
- Robust against unexpected system failure using GNU make
- easy restart after failure
- Massively parallel, can run hundreds of jobs
- Splits large jobs into many pieces
- Simplifies running on clusters
- Scalable to tens of thousands of samples
- Easy to use - Automates series of configurable steps
- user doesn't have to understand/configure/know the many tools required to create high quality results
- Available on Amazon Web Services (AWS) Elastic Compute Cloud (EC2)
- Run on local machines/clusters
- Available via Debian Packages
GotCloud incorporates the alignment and variant calling pipelines that we have been using at UM into one easy to use, publicly available tool. GotCloud can run on a user's computer, on an instance in a compute cloud, and/or can split the work up onto a cluster of machines or instances.
Join GotCloud mailing list
Please join in the GotCloud Google Group to ask / discuss / comment about these pipelines.
Currently the "join" button appears to be missing. Click "NEW TOPIC", then select "Join this group". You can then cancel the message post (or post a message).
You can also email Mary Kate Wing (mktrost@umich.edu).
Sequence Analysis Background Information
There are many essential steps in the analysis of next generation sequence data.
Next generation sequence data analysis starts with FASTQ files, the typical format provided from your sequencing center containing the sequence & base quality information for your data.
The fastq files are processed using the alignment pipeline which finds the most likely genomic location for each read and stores that information in a BAM (Binary Sequence Alignment/Map format) file. In addition to the sequence and base quality information contained in FASTQ files, a BAM file also contains the genomic location and some additional information about the mapping. As part of the alignment pipeline, the base qualities are adjusted to more accurately reflect the likelihood that the base is correct.
The alignment pipeline can be skipped if you already have Deduped and Recalibrated BAM files.
The variant calling pipeline processes the deduped and recalibrated BAMs file produced by the alignment pipeline or that you provide it, generating an initial list of polymorphic sites and genotypes stored in a VCF (Variant Call Format) file. The variant calling pipeline then filters the variants using both hard and a Support Vector Machine (SVM). It then uses haplotype information to refine these genotypes in an updated VCF file.
After completing the GotCloud Variant Calling PIpeline, EPACTS (Efficient and Parallelizable Association Container Toolbox) can be used to perform statistical tests to identify genome-wide association from sequence data.
GotCloud Setup
You may run the GotCloud software in several modes:
- On your own hardware running Ubuntu or Redhat/CentOS. See the instructions about installing the software below.
- On any EC2 instance that uses Ubuntu or Redhat/CentOS distribution. You can install the software as described below, or create a volume using our snapshot (see Amazon Snapshot).
- On an EC2 cluster instance created by StarCluster. You can install the software as described below, or create a volume using our snapshot (see Amazon Snapshot).
Details for the Choices of Your Install
AWS
The following describes the use of this software with the Amazon Web Services (https://aws.amazon.com/), but you can just as easily use the pipelines on your own machine(s) by just installing them.
Latest Documentation at Tutorial: GotCloud
Install GotCloud Software
- As a debian package
- As a red hat package
- As a tar
- On Amazon EC2
- Create a volume using our Amazon Snapshot
- Create an EC2 cluster instance using StarCluster
Install Resource Files
- Install genetic resource files
Resources / Cost
Configure
- Configure Gotcloud for your installation
Running GotCloud Software
Development Notes
- Creating Packages
- Amazon EC2
- Creating a Snapshot on EC2
- Creating an AMI on EC2
- Mount S3 Volume
- Notes on sequence data preparation in Amazon Storage.