Difference between revisions of "GREGOR"
| (43 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | == | + | ==GREGOR== |
| − | ''' | + | '''GREGOR''' ('''G'''enomic '''R'''egulatory '''E'''lements and '''G'''was '''O'''verlap algo'''R'''ithm) is a tool built to evaluate global enrichment of trait-associated variants in experimentally annotated epigenomic regulatory features. |
| − | + | Because all reference data are version hg19, please make sure that your index SNP list and BED files are also version hg19. | |
| − | == | + | == Get GREGOR Source Codes == |
| − | Through this link [http:// | + | === Download from webpage === |
| + | Through this link [http://csg.sph.umich.edu/GREGOR/ GREGOR], you can download a copy of GREGOR. | ||
| − | === | + | == Build GREGOR == |
| − | |||
| − | + | To build GREGOR, copy the GREGOR package to the directory you want, and then run the following command: | |
| − | |||
| − | |||
| − | + | tar xzvf GREGOR.v1.4.0.tar.gz | |
| − | + | After you unzip, in the folder "GREGOR" you can find 4 directories (./Copyrights, ./example, ./lib ./script) and 2 files (README, release_version.txt). | |
| − | |||
| − | |||
| − | |||
| − | == | + | == Download reference files == |
| + | ownload the reference files from this link [http://csg.sph.umich.edu/GREGOR/ GREGOR Download]. | ||
| − | + | Reference files are created for the different population groups(AFR, AMR, ASN, EUR, SAN) from 1000G data (Release date : May 21, 2011). | |
| − | + | If your LD r2 threshold equals or greater than 0.7, please download reference files from category: LD window size = 1MB; LD r2 ≥ 0.7. | |
| + | If your LD r2 threshold equals or greater than 0.2, please download reference files from category: LD window size = 1MB; LD r2 ≥ 0.2. | ||
| − | After | + | After download reference files, you need merge the part files to one gz file. Use the command line likes: |
| − | + | cat \ | |
| + | GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.00 \ | ||
| + | GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.01 \ | ||
| + | GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.02 \ | ||
| + | GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.03 \ | ||
| + | GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.04 \ | ||
| + | > GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz | ||
| + | |||
| + | Then extract this file: | ||
| + | tar zxvf GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz | ||
| − | + | You will get one directory which has the name "AFR". | |
| − | |||
| − | |||
| − | |||
| − | |||
== Basic Usage Example == | == Basic Usage Example == | ||
| Line 43: | Line 45: | ||
When you run | When you run | ||
| − | perl | + | perl GREGOR.pl |
| − | you will get some information about | + | you will get some information about GREGOR |
---------------------------------------------------------------------------------- | ---------------------------------------------------------------------------------- | ||
| − | + | GREGOR.pl : Functional annotation of trait-associated variants | |
---------------------------------------------------------------------------------- | ---------------------------------------------------------------------------------- | ||
This program tests for enrichment of an input list of trait-associated index | This program tests for enrichment of an input list of trait-associated index | ||
| Line 61: | Line 63: | ||
Report Bug(s) : jich[at]umich[dot]edu | Report Bug(s) : jich[at]umich[dot]edu | ||
---------------------------------------------------------------------------------- | ---------------------------------------------------------------------------------- | ||
| − | Usage : perl | + | Usage : perl GREGOR.pl --conf [conf.file] |
---------------------------------------------------------------------------------- | ---------------------------------------------------------------------------------- | ||
| Line 67: | Line 69: | ||
The following command is a typical command line: | The following command is a typical command line: | ||
| − | perl | + | perl GREGOR.pl --conf [conf.file] |
Example configuration file can be found in example directory. Users have to modify the configurations before running. | Example configuration file can be found in example directory. Users have to modify the configurations before running. | ||
== Configuration File == | == Configuration File == | ||
| − | The example configuration file below illustrate how to configure the | + | The example configuration file below illustrate how to configure the GREGOR configuration file. |
############################################################################### | ############################################################################### | ||
| Line 81: | Line 83: | ||
## KEY ELEMENTS TO CONFIGURE : NEED TO MODIFY | ## KEY ELEMENTS TO CONFIGURE : NEED TO MODIFY | ||
############################################################################### | ############################################################################### | ||
| − | INDEX_SNP_FILE = / | + | INDEX_SNP_FILE = /workingdirectory/example/example.index.snps.rsid.list.txt |
| − | BED_FILE_INDEX = / | + | BED_FILE_INDEX = /workingdirectory/example/example.bed.file.index |
| + | REF_DIR = /workingdirectory/ref/ | ||
R2THRESHOLD = 0.7 | R2THRESHOLD = 0.7 | ||
LDWINDOWSIZE = 1000000 | LDWINDOWSIZE = 1000000 | ||
| − | OUT_DIR = / | + | OUT_DIR = /workingdirectory/example/example.rsid.20130808/ |
MIN_NEIGHBOR_NUM = 500 | MIN_NEIGHBOR_NUM = 500 | ||
BEDFILE_IS_SORTED = True | BEDFILE_IS_SORTED = True | ||
| − | + | POPULATION = AFR ## define the population, you can specify EUR, AFR, AMR or ASN | |
| + | TOPNBEDFILES = 2 | ||
| + | JOBNUMBER = 10 | ||
| + | ############################################################################### | ||
| + | #BATCHTYPE = mosix ## submit jobs on MOSIX | ||
| + | #BATCHOPTS = -E/tmp -i -m2000 -j10,11,12,13,14,15,16,17,18,19,120,122,123,124,125 sh -c | ||
| + | ############################################################################### | ||
| + | #BATCHTYPE = slurm ## submit jobs on SLURM | ||
| + | #BATCHOPTS = --partition=main --time=0:30:0 | ||
| + | ############################################################################### | ||
| + | BATCHTYPE = local ## run jobs on local machine | ||
| + | |||
In the config file, there are several parameters to adjust: | In the config file, there are several parameters to adjust: | ||
| Line 96: | Line 110: | ||
BED_FILE_INDEX: This file lists the datasets (e.g. BED files) to be used for enrichment analysis. Use complete paths to file locations and make sure positions are in hg19 format. | BED_FILE_INDEX: This file lists the datasets (e.g. BED files) to be used for enrichment analysis. Use complete paths to file locations and make sure positions are in hg19 format. | ||
| − | R2THRESHOLD and LDWINDOWSIZE: These two parameters define the index SNP LD proxies by | + | REF_DIR: Define reference file directory which you download at here. If your "AFR" folder is at "/home/myid/GRGORE/ref/AFR/", then define this parameter to "/home/myid/GRGORE/ref/". |
| + | |||
| + | R2THRESHOLD and LDWINDOWSIZE: These two parameters define the index SNP (and control SNP) LD proxies by r2 threshold and LD window size. If you download r2 ≥ 0.7, you can define this number between 1 and 0.7. | ||
OUT_DIR: All result files are saved to this folder, where the script will create multiple sub-directories. Index SNPs are in the folder "index_SNP"; Random SNPs are in the folder "random_SNP". | OUT_DIR: All result files are saved to this folder, where the script will create multiple sub-directories. Index SNPs are in the folder "index_SNP"; Random SNPs are in the folder "random_SNP". | ||
| − | MIN_NEIGHBOR_NUM: Define | + | MIN_NEIGHBOR_NUM: Define the minimum number of control SNPs for each index SNP. Script will find no less than this number around every index SNP. If you make this number of control SNPs very large, the control SNPs will be less closely matched on the three matching properties (distance to nearest gene, frequency and number of SNPs in LD). |
BEDFILE_IS_SORTED: True or false, depending on whether the BED files listed in the index file are sorted. | BEDFILE_IS_SORTED: True or false, depending on whether the BED files listed in the index file are sorted. | ||
| + | |||
| + | POPULATION: If you use reference file "AFR", define this to AFR. You have 5 optiones: AFR, AMR, ASN, EUR and SAN. | ||
| + | |||
| + | GREGOR can run on local machine or on the cluster with MOSIX or SLURM. | ||
| + | BATCHTYPE: When you run GREGOR on local machine, specify "local"; when run on MOSIX system, specify "mosix"; when run on SLURM system, specify "slurm". | ||
| + | |||
| + | BATCHOPTS: This parameter works with BATCHTYPE when you specify "mosix" or "slurm". For example, when you define mosix, this parameter can be "-E/tmp -i -m2000 -j10,11,12,13,14,15,16 sh -c"; when you define "slurm", it can be "--partition=1000g --time=0:30:0" | ||
| + | |||
| + | == Reference Files == | ||
| + | We provide two kinds of reference files. The difference between these reference data are LD buddy definitions. | ||
| + | *LD window size = 1MB; LD r2 ≥ 0.7: | ||
| + | **All LD buddies are in window size 1MB and r2 is greater than and equals to 0.7. If you want to calculate LD buddies in 1MB and r2 ≥ 0.7 (such as 0.9,0.8,0.7), please use these reference data. | ||
| + | *LD window size = 1MB; LD r2 ≥ 0.2: | ||
| + | **All LD buddies are in window size 1MB and r2 is greater than and equals to 0.2. If you want to calculate LD buddies in 1MB and r2 ≥ 0.2 (such as 0.6,0.5,0.4,0.3,0.2), please use these reference data. | ||
== Results Output == | == Results Output == | ||
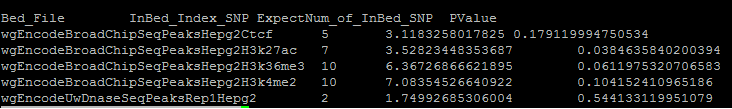
The file StatisticSummaryFile.txt in the output directory contains enrichment results with the following information: | The file StatisticSummaryFile.txt in the output directory contains enrichment results with the following information: | ||
| + | |||
| + | [[File:GREGOR Summary 20160201.png]] | ||
Bed_File: The individual datasets used in the enrichment analysis | Bed_File: The individual datasets used in the enrichment analysis | ||
| Line 113: | Line 145: | ||
Pvalue: P-value calculated assuming a sum of binomial distributions to represent the number of index SNPs (or LD proxies) that overlap a dataset compared to the expectation observed in the matched control sets | Pvalue: P-value calculated assuming a sum of binomial distributions to represent the number of index SNPs (or LD proxies) that overlap a dataset compared to the expectation observed in the matched control sets | ||
| − | *Note: SNPs that cannot be converted from rsID to chr:pos format are listed in the output file rsid.index.snp.txt. SNPs for which there are no LD proxies or no MAF data available are listed in the output file nonannoted.index.snp.txt. | + | *Note: |
| + | **SNPs that cannot be converted from rsID to chr:pos format are listed in the output file rsid.index.snp.txt. SNPs for which there are no LD proxies or no MAF data available are listed in the output file nonannoted.index.snp.txt. | ||
| + | ** If one index SNP and its LD-buddies are not in any bed region, the Pvalue could be defined to "NA" | ||
| + | |||
| + | == Testing GREGOR == | ||
| + | There is an example directory in ~/GREGOR. You can find index SNP file, 3 bed files, bed file index and example config file. | ||
| + | After change your config file, you can run a test. | ||
| + | |||
| + | perl ~/GREGOR/script/GREGOR.pl --conf ~/GREGOR/example/example.conf | ||
| + | |||
| + | After running 2 minutes +/- 1 minutes. You will get result file "StatisticSummaryFile.txt" in your defined output directory. | ||
== Acknowledgements == | == Acknowledgements == | ||
| − | + | GREGOR is the result of collaborative efforts by Cristen Willer, Jin Chen, Wei Zhou, Ellen Schmidt, He Zhang, and Goncalo Abecasis. Please email Cristen Willer [cristen@umich.edu] with any questions. | |
Latest revision as of 17:39, 15 November 2016
GREGOR
GREGOR (Genomic Regulatory Elements and Gwas Overlap algoRithm) is a tool built to evaluate global enrichment of trait-associated variants in experimentally annotated epigenomic regulatory features.
Because all reference data are version hg19, please make sure that your index SNP list and BED files are also version hg19.
Get GREGOR Source Codes
Download from webpage
Through this link GREGOR, you can download a copy of GREGOR.
Build GREGOR
To build GREGOR, copy the GREGOR package to the directory you want, and then run the following command:
tar xzvf GREGOR.v1.4.0.tar.gz
After you unzip, in the folder "GREGOR" you can find 4 directories (./Copyrights, ./example, ./lib ./script) and 2 files (README, release_version.txt).
Download reference files
ownload the reference files from this link GREGOR Download.
Reference files are created for the different population groups(AFR, AMR, ASN, EUR, SAN) from 1000G data (Release date : May 21, 2011).
If your LD r2 threshold equals or greater than 0.7, please download reference files from category: LD window size = 1MB; LD r2 ≥ 0.7. If your LD r2 threshold equals or greater than 0.2, please download reference files from category: LD window size = 1MB; LD r2 ≥ 0.2.
After download reference files, you need merge the part files to one gz file. Use the command line likes:
cat \ GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.00 \ GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.01 \ GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.02 \ GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.03 \ GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz.part.04 \ > GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz
Then extract this file:
tar zxvf GREGOR.AFR.ref.r2.greater.than.0.2.tar.gz
You will get one directory which has the name "AFR".
Basic Usage Example
When you run
perl GREGOR.pl
you will get some information about GREGOR
GREGOR.pl : Functional annotation of trait-associated variants
This program tests for enrichment of an input list of trait-associated index SNPs ([chr:pos] format or rsID, hg19) in experimentally annotated regulatory domains (BED files).
Note: the index SNPs should be hg19 version. All maf and LD data are from 1000G EUR samples! (Release date : May 21, 2011)
Version : 1.1.0
Report Bug(s) : jich[at]umich[dot]edu
Usage : perl GREGOR.pl --conf [conf.file]
The following command is a typical command line:
perl GREGOR.pl --conf [conf.file]
Example configuration file can be found in example directory. Users have to modify the configurations before running.
Configuration File
The example configuration file below illustrate how to configure the GREGOR configuration file.
############################################################################### # CHIPSEQ ENRICHMENT CONFIGURATION FILE # This configuration file contains run-time configuration of # CHIP_SEQ ENRICHMENT ############################################################################### ## KEY ELEMENTS TO CONFIGURE : NEED TO MODIFY ############################################################################### INDEX_SNP_FILE = /workingdirectory/example/example.index.snps.rsid.list.txt BED_FILE_INDEX = /workingdirectory/example/example.bed.file.index REF_DIR = /workingdirectory/ref/ R2THRESHOLD = 0.7 LDWINDOWSIZE = 1000000 OUT_DIR = /workingdirectory/example/example.rsid.20130808/ MIN_NEIGHBOR_NUM = 500 BEDFILE_IS_SORTED = True POPULATION = AFR ## define the population, you can specify EUR, AFR, AMR or ASN TOPNBEDFILES = 2 JOBNUMBER = 10 ############################################################################### #BATCHTYPE = mosix ## submit jobs on MOSIX #BATCHOPTS = -E/tmp -i -m2000 -j10,11,12,13,14,15,16,17,18,19,120,122,123,124,125 sh -c ############################################################################### #BATCHTYPE = slurm ## submit jobs on SLURM #BATCHOPTS = --partition=main --time=0:30:0 ############################################################################### BATCHTYPE = local ## run jobs on local machine
In the config file, there are several parameters to adjust:
INDEX_SNP_FILE: This file contains a single column of trait-associated input SNPs, without a header. Variants can be listed in rsid or hg19 chr:pos format.
BED_FILE_INDEX: This file lists the datasets (e.g. BED files) to be used for enrichment analysis. Use complete paths to file locations and make sure positions are in hg19 format.
REF_DIR: Define reference file directory which you download at here. If your "AFR" folder is at "/home/myid/GRGORE/ref/AFR/", then define this parameter to "/home/myid/GRGORE/ref/".
R2THRESHOLD and LDWINDOWSIZE: These two parameters define the index SNP (and control SNP) LD proxies by r2 threshold and LD window size. If you download r2 ≥ 0.7, you can define this number between 1 and 0.7.
OUT_DIR: All result files are saved to this folder, where the script will create multiple sub-directories. Index SNPs are in the folder "index_SNP"; Random SNPs are in the folder "random_SNP".
MIN_NEIGHBOR_NUM: Define the minimum number of control SNPs for each index SNP. Script will find no less than this number around every index SNP. If you make this number of control SNPs very large, the control SNPs will be less closely matched on the three matching properties (distance to nearest gene, frequency and number of SNPs in LD).
BEDFILE_IS_SORTED: True or false, depending on whether the BED files listed in the index file are sorted.
POPULATION: If you use reference file "AFR", define this to AFR. You have 5 optiones: AFR, AMR, ASN, EUR and SAN.
GREGOR can run on local machine or on the cluster with MOSIX or SLURM. BATCHTYPE: When you run GREGOR on local machine, specify "local"; when run on MOSIX system, specify "mosix"; when run on SLURM system, specify "slurm".
BATCHOPTS: This parameter works with BATCHTYPE when you specify "mosix" or "slurm". For example, when you define mosix, this parameter can be "-E/tmp -i -m2000 -j10,11,12,13,14,15,16 sh -c"; when you define "slurm", it can be "--partition=1000g --time=0:30:0"
Reference Files
We provide two kinds of reference files. The difference between these reference data are LD buddy definitions.
- LD window size = 1MB; LD r2 ≥ 0.7:
- All LD buddies are in window size 1MB and r2 is greater than and equals to 0.7. If you want to calculate LD buddies in 1MB and r2 ≥ 0.7 (such as 0.9,0.8,0.7), please use these reference data.
- LD window size = 1MB; LD r2 ≥ 0.2:
- All LD buddies are in window size 1MB and r2 is greater than and equals to 0.2. If you want to calculate LD buddies in 1MB and r2 ≥ 0.2 (such as 0.6,0.5,0.4,0.3,0.2), please use these reference data.
Results Output
The file StatisticSummaryFile.txt in the output directory contains enrichment results with the following information:
Bed_File: The individual datasets used in the enrichment analysis
InBed_Index_SNP: Number of index SNPs or their LD proxies that overlaps regulatory regions in each dataset
Pvalue: P-value calculated assuming a sum of binomial distributions to represent the number of index SNPs (or LD proxies) that overlap a dataset compared to the expectation observed in the matched control sets
- Note:
- SNPs that cannot be converted from rsID to chr:pos format are listed in the output file rsid.index.snp.txt. SNPs for which there are no LD proxies or no MAF data available are listed in the output file nonannoted.index.snp.txt.
- If one index SNP and its LD-buddies are not in any bed region, the Pvalue could be defined to "NA"
Testing GREGOR
There is an example directory in ~/GREGOR. You can find index SNP file, 3 bed files, bed file index and example config file. After change your config file, you can run a test.
perl ~/GREGOR/script/GREGOR.pl --conf ~/GREGOR/example/example.conf
After running 2 minutes +/- 1 minutes. You will get result file "StatisticSummaryFile.txt" in your defined output directory.
Acknowledgements
GREGOR is the result of collaborative efforts by Cristen Willer, Jin Chen, Wei Zhou, Ellen Schmidt, He Zhang, and Goncalo Abecasis. Please email Cristen Willer [cristen@umich.edu] with any questions.