Difference between revisions of "GotCloud: Alignment Pipeline"
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
Back to parent: [[GotCloud]] | Back to parent: [[GotCloud]] | ||
| − | The Alignment/Mapping Pipeline takes FASTQ files and generates recalibrated BAM files from them. | + | |
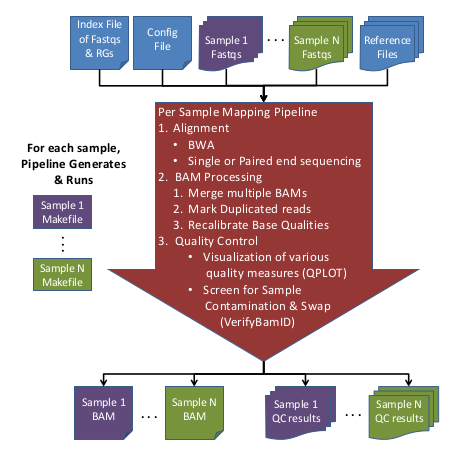
| + | == Overview of Alignment Pipeline Steps == | ||
| + | The Alignment/Mapping Pipeline takes [http://en.wikipedia.org/wiki/FASTQ_format FASTQ files] and generates recalibrated [[BAM|BAM (Binary Sequence Alignment/Map format) files]] from them. | ||
| + | |||
| + | [[File:MappingSteps.png]] | ||
== Running the GotCloud Alignment Pipeline == | == Running the GotCloud Alignment Pipeline == | ||
The alignment pipeline is run using the <code>align</code> option of the <code>gotcloud</code> script. This option calls <code>align.pl</code> found in the <code>bin/</code> directory under the <code>gotcloud</code> installation. | The alignment pipeline is run using the <code>align</code> option of the <code>gotcloud</code> script. This option calls <code>align.pl</code> found in the <code>bin/</code> directory under the <code>gotcloud</code> installation. | ||
| + | |||
| + | Use the <code>--conf</code> parameter followed by the configuration file to specify the configuration to use for this run of the alignment pipeline. | ||
| + | |||
| + | You must specify the input list of FASTQs mapped to sample id to tell the alignment pipeline what files to process. You can do this by setting either: | ||
| + | * <code>FASTQ_LIST</code> in the configuration file | ||
| + | * <code>--list</code> on the command-line | ||
| + | |||
| + | You must specify an output directory to tell the alignment pipeline where to write its output by either setting: | ||
| + | * <code>OUT_DIR</code> in the configuration file | ||
| + | * <code>--outdir</code> on the command-line | ||
| + | |||
| + | '''Example of a Basic Alignment Command''' | ||
| + | gotcloud align --conf myAlignTest.conf --outdir ~/gotcloudOutput/align/ | ||
| + | |||
===Running the Automated Test=== | ===Running the Automated Test=== | ||
| Line 17: | Line 34: | ||
where OUTPUT_DIR is the directory where you want to store the test results | where OUTPUT_DIR is the directory where you want to store the test results | ||
| − | If you see "Successfully ran the test case, congratulations!", then you are ready to align samples. | + | If you see "Successfully ran the test case, congratulations!", then you are ready to align samples. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Input Data:== | == Input Data:== | ||
*Raw Sequence (FASTQ) files | *Raw Sequence (FASTQ) files | ||
| − | * | + | *FASTQ List file mapping fastq pairs to sample (optional: Read Group information) |
*Reference files | *Reference files | ||
*(Optional) Configuration file to override default options | *(Optional) Configuration file to override default options | ||
| Line 35: | Line 46: | ||
These are the FASTQ files that need to be mapped to BAM files. | These are the FASTQ files that need to be mapped to BAM files. | ||
| − | These files are specified in the [[# | + | These files are specified in the [[#FASTQ List File|FASTQ List File]]. |
| − | === | + | === FASTQ List File === |
| − | This file specifies the FASTQ files that need to be processed | + | This file specifies the FASTQ files that need to be processed. It maps the FASTQ pairs to the associated Sample ID. Optionally Read Group information for the FASTQ pairs can be specified. If the Read Group information is not specified, it is inferred. |
| − | This file is specified either via the command line parameter <code>-- | + | This file is specified either via the command line parameter <code>--list</code> or via the configuration file setting <code>FASTQ_LIST</code>. |
The command-line setting takes precedence over the configuration file setting. | The command-line setting takes precedence over the configuration file setting. | ||
| − | The | + | The FASTQ list is a tab delimited file that starts with a header line. The columns may be in any order. |
Following the header line, there is one line per single-end read and one line per paired-end read (only 1 line per pair). | Following the header line, there is one line per single-end read and one line per paired-end read (only 1 line per pair). | ||
Required Column Names: | Required Column Names: | ||
| − | * MERGE_NAME - base name for the resulting BAM file for the sample (used to group multiple fastqs or fastq pairs into a single BAM) | + | * MERGE_NAME - base name for the resulting BAM file for the sample (used to group multiple fastqs or fastq pairs into a single BAM) |
| + | ** The SAMPLE column can be specified instead of MERGE_NAME. SAMPLE will be used for both the sample and the base name. | ||
* FASTQ1 - name of the fastq or the first in the pair if paired-end. (Only 1 line per pair) | * FASTQ1 - name of the fastq or the first in the pair if paired-end. (Only 1 line per pair) | ||
Optional Column Names: | Optional Column Names: | ||
* FASTQ2 - name of the 2nd fastq in paired-end reads. Specify '.' if the column exists, but this line is single-ended. | * FASTQ2 - name of the 2nd fastq in paired-end reads. Specify '.' if the column exists, but this line is single-ended. | ||
| − | * RGID - Read Group ID for this entry | + | * RGID - Read Group ID for this entry |
| + | ** If this field is not specified, the first line of the fastq will be used to determine the RG. | ||
| + | *** If the first line does not match the expected format for determining RG, incrementing numbers per fastq file will be used. | ||
* SAMPLE - Sample Name for this entry | * SAMPLE - Sample Name for this entry | ||
| + | ** If SAMPLE is not specified, MERGE_NAME will be used for the sample name | ||
* LIBRARY - Library for this entry | * LIBRARY - Library for this entry | ||
| + | ** If LIBRARY is not specified, the sample name will be used | ||
* CENTER - Center Name for this entry | * CENTER - Center Name for this entry | ||
| + | ** If CENTER is not specified, it will default to "unknown" | ||
* PLATFORM - Platform for this entry | * PLATFORM - Platform for this entry | ||
| + | ** If PLATFORM is not specified, it will default to ILLUMINA | ||
| − | The RGID, SAMPLE, LIBRARY, CENTER, and PLATFORM are used to populate the Read Group information for this entry. | + | The RGID, SAMPLE, LIBRARY, CENTER, and PLATFORM are used to populate the Read Group information for this entry. |
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM | MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM | ||
| Line 68: | Line 86: | ||
Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA | Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA | ||
| − | The <code>-- | + | The <code>--fastq_prefix</code>/<code>FASTQ_PREFIX</code> setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files. |
=== Reference Files === | === Reference Files === | ||
| + | See [[GotCloud: Genetic Reference and Resource Files]] for detailed information about the multiple required reference files for the alignment pipeline, including: | ||
| + | * How to obtain default references | ||
| + | * Configuration keys & default values | ||
| + | * How to generate your own references | ||
| + | * How to point GotCloud to your reference files | ||
| − | + | Required Reference File Types: | |
| − | * Reference | + | * [[GotCloud: Genetic Reference and Resource Files#Reference fasta Files|Reference fasta Files]] |
| − | + | * [[GotCloud: Genetic Reference and Resource Files#DBSNP VCF Files|DBSNP VCF Files]] | |
| − | + | * [[GotCloud: Genetic Reference and Resource Files#HapMap3 VCF Files|HapMap3 VCF Files]] | |
| − | * | ||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Configuration File === | === Configuration File === | ||
| − | Configuration | + | {{:GotCloud: Configuration}} |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==== Recommended Settings ==== | |
| − | + | As of GotCloud version 1.16, the alignment pipeline uses <code>bwa mem</code> by default. Prior to version 1.16, the default aligner was <code>bwa aln</code>. | |
| − | + | You can override the defaults by setting in your configuration file: | |
| + | * to use <code>bwa mem</code> (you do not need to set this in version 1.16 and later since it is the default) | ||
| + | MAP_TYPE = BWA_MEM | ||
| + | * to use <code>bwa aln</code> (you do not need to set this prior to version 1.16 since it is the default) | ||
| + | MAP_TYPE = BWA | ||
| − | ==== Required Settings ==== | + | ==== Additional Required Settings ==== |
| − | |||
| − | See [[# | + | See [[#FASTQ List File|FASTQ List File]] for how to set the index file either via command line options or via configuration. |
==== Turning Off Optional Steps==== | ==== Turning Off Optional Steps==== | ||
Quality Control steps can be disabled. | Quality Control steps can be disabled. | ||
| − | To Disable QPLOT, | + | To Disable QPLOT, remove qplot from the PER_MERGE_STEPS configuration by setting: |
| − | + | PER_MERGE_STEPS = verifyBamID index recab | |
| − | To Disable VerifyBamID, | + | |
| − | + | To Disable VerifyBamID, remove qplot from the PER_MERGE_STEPS configuration by setting: | |
| + | PER_MERGE_STEPS = qplot index recab | ||
==== Optional Configurable Settings ==== | ==== Optional Configurable Settings ==== | ||
| Line 121: | Line 133: | ||
* BWA_THREADS = -t N | * BWA_THREADS = -t N | ||
| − | ** Fill in the N with the number of threads you want BWA to run with, default is 1 | + | ** Fill in the N with the number of threads you want BWA to run with, default is 1 |
| − | * | + | * BWA_QUAL = -q N |
| − | ** | + | ** Fill in the N with the trim quality you want BWA aln to run with, default is 15. This parameter is only applied to bwa aln. It is not used for BWA_MEM. |
| − | * | + | * BWA_MEM_OPTS = |
| − | ** | + | ** Specify any additional bwa mem options using this parameter. |
| − | * | + | * SORT_MAX_MEM = 2000000000 |
| − | ** | + | ** Maximum amount of memory used by samtools sort after running bwa |
== Running the Alignment Pipeline == | == Running the Alignment Pipeline == | ||
| Line 134: | Line 146: | ||
* help - print usage | * help - print usage | ||
* test OUTPUT_DIR - run the test example placing the output in a user specified OUTPUT_DIR. No other options are required. | * test OUTPUT_DIR - run the test example placing the output in a user specified OUTPUT_DIR. No other options are required. | ||
| − | * | + | * outdir OUTPUT_DIR - directory for the output |
** May also be specified via OUT_DIR in the configuration file | ** May also be specified via OUT_DIR in the configuration file | ||
** Required to be set either via command-line or configuration | ** Required to be set either via command-line or configuration | ||
* conf CONFIG_FILE - configuration file | * conf CONFIG_FILE - configuration file | ||
| − | * | + | * list FASTQ_LIST_FILE_NAME - name of the fastq list file |
| − | ** May also be specified via | + | ** May also be specified via FASTQ_LIST in the configuration file |
** Required to be set either via command-line or configuration | ** Required to be set either via command-line or configuration | ||
* ref_dir REFERENCE_DIR - value to set config key REF_DIR to, overriding other values, REF_DIR can then be used inside config files. | * ref_dir REFERENCE_DIR - value to set config key REF_DIR to, overriding other values, REF_DIR can then be used inside config files. | ||
** May also be specified via REF_DIR in the configuration file | ** May also be specified via REF_DIR in the configuration file | ||
| − | * | + | * ref_prefix REFERENCE_DIR - path to prepend to non-absolute REF paths. |
| − | ** May also be specified via | + | ** May also be specified via REF_PREFIX in the configuration file |
| + | * fastq_prefix FASTQ_PATH - prefix path to the fastq files specified in the FASTQ_LIST | ||
| + | ** May also be specified via FASTQ_PREFIX in the configuration file | ||
| + | * base_prefix BASE_PATH - prefix path to the prepend to fastq/ref files without absolute paths | ||
| + | ** May also be specified via BASE_PREFIX in the configuration file | ||
* keepTmp - Do not remove the temporary files (removed by default) | * keepTmp - Do not remove the temporary files (removed by default) | ||
** May also be specified via KEEP_TMP in the configuration file | ** May also be specified via KEEP_TMP in the configuration file | ||
| − | * | + | * keepLog - Do not remove the intermediate log files (removed by default) |
| − | * | + | ** May also be specified via KEEP_LOG in the configuration file |
| − | + | * numjobs N - Replace N with the number of samples that should be processed in parallel | |
| + | * threads N - Replace N with the number of targets in each makefile that should be run in parallel | ||
| + | * dryrun - Create the Makefile, but do not run it | ||
| + | * maxlocaljobs N - Replace N with the maximum number of jobs that can be run locally (no batchtype specified). Default is 10. | ||
| + | * batchtype TYPE - Tells GotCloud the specified batch type to send jobs to the client nodes | ||
| + | ** May also be specified via BATCH_TYPE in the configuration file | ||
| + | ** Can be: mosix, slurm, slurmi, pbs, sge, sgei | ||
| + | * batchopts OPTS - Tells GotCloud the options to pass onto the batch system | ||
| + | ** May also be specified via BATCH_OPTS in the configuration file | ||
| + | * noPhoneHome - disable the phone home logic | ||
| + | * gotcloudroot DIR - Specifies an alternate path to other gotcloud files rather than using the path to the gotcloud/align.pl. | ||
Note: Command-line options take priority over configuration file settings | Note: Command-line options take priority over configuration file settings | ||
| Line 165: | Line 191: | ||
On success, you will see: | On success, you will see: | ||
| − | Processing finished in nn secs with no errors reported | + | Processing finished in nn secs with no errors reported |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | You should see a <code>.OK</code> for each Sample in the index file. | + | If processing fails part way through, you can pick up where you left off by rerunning gotcloud or the make command. |
| + | |||
| + | === Alignment Pipeline Output === | ||
| + | Upon successful completion of the alignment pipeline, you should see the following files/ subdirectories under the user specified output directory: | ||
| + | * '''bam.list''' - file containing sample->BAM mapping that can be used in other GotCloud pipelines | ||
| + | * '''bams/''' - contains the final BAM and bai (BAM index) files | ||
| + | ** '''*.recal.bam''' | ||
| + | ** '''*.recal.bam.bai''' | ||
| + | ** ''*.recal.bam.bai.done'' - temp file indicating this step completed successfully | ||
| + | ** ''*.recal.bam.done'' - temp file indicating this step completed successfully | ||
| + | ** *.recal.bam.metrics - dedup & recalibration log | ||
| + | ** *.recal.bam.qemp - recalibration tables | ||
| + | * Makefiles/ - contains the Makefiles and logs used by GotCloud to run the alignment pipeline | ||
| + | * '''QCFiles/''' - contains quality control results if quality control is not disabled | ||
| + | ** VerifyBamID Output - see [[VerifyBamID#A_guideline_to_interpret_output_files|VerifyBamID: A guideline to interpret output files]] for more information | ||
| + | *** *.genoCheck.depthRG - depth distribution of the sequence reads per read group | ||
| + | *** *.genoCheck.depthSM - depth distribution of the sequence reads per sample | ||
| + | *** ''*.genoCheck.done'' - temp file indicating this step completed successfully | ||
| + | *** *.genoCheck.selfRG - per-readGroup statistics describing how well each lane matches to the annotated sample | ||
| + | *** '''*.genoCheck.selfSM''' - main output file containing the contamination estimate; per-sample statistics describing how well the sample matches to the annotated sample | ||
| + | **** Check the 'FREEMIX' column for genotype-free estimate of contamination 0-1 scale, the lower, the better | ||
| + | **** If [FREEMIX] >= 0.03 and [FREELK1]-[FREELK0] is large, possible contamination | ||
| + | ** Qplot Output - see: [[QPLOT#Diagnose_sequencing_quality|QPLOT: Diagnose sequencing quality]] for more info on how to use QPLOT results | ||
| + | *** ''*.qplot.done'' - temp file indicating this step completed successfully | ||
| + | *** '''*.qplot.R''' - Rscript that can be used to generate the pdf graphs | ||
| + | *** '''*.qplot.stats''' - sample statistics | ||
| + | * tmp/ - contains intermediate files (most are deleted unless --keepTmp is specified) | ||
| + | * *.OK - one OK file per sample; indicates the Sample successfully completed alignment | ||
| + | |||
| + | You should also see a <code>.OK</code> for each Sample in the index file. | ||
If you do not see these <code>.OK</code> files, then your Alignment Pipeline failed. | If you do not see these <code>.OK</code> files, then your Alignment Pipeline failed. | ||
| − | On success, the bams/ directory contains the final BAMs and bais. | + | '''On success, the bams/ directory contains the final BAMs and bais.''' |
| − | |||
| − | |||
Latest revision as of 18:39, 28 January 2016
Back to parent: GotCloud
Overview of Alignment Pipeline Steps
The Alignment/Mapping Pipeline takes FASTQ files and generates recalibrated BAM (Binary Sequence Alignment/Map format) files from them.
Running the GotCloud Alignment Pipeline
The alignment pipeline is run using the align option of the gotcloud script. This option calls align.pl found in the bin/ directory under the gotcloud installation.
Use the --conf parameter followed by the configuration file to specify the configuration to use for this run of the alignment pipeline.
You must specify the input list of FASTQs mapped to sample id to tell the alignment pipeline what files to process. You can do this by setting either:
FASTQ_LISTin the configuration file--liston the command-line
You must specify an output directory to tell the alignment pipeline where to write its output by either setting:
OUT_DIRin the configuration file--outdiron the command-line
Example of a Basic Alignment Command
gotcloud align --conf myAlignTest.conf --outdir ~/gotcloudOutput/align/
Running the Automated Test
The automated test runs the alignment pipeline on a small set of test data and checks that the results against expected results validating that GotCloud is installed correctly.
- Run alignment pipeline test:
gotcloud align --test OUTPUT_DIR
where OUTPUT_DIR is the directory where you want to store the test results
If you see "Successfully ran the test case, congratulations!", then you are ready to align samples.
Input Data:
- Raw Sequence (FASTQ) files
- FASTQ List file mapping fastq pairs to sample (optional: Read Group information)
- Reference files
- (Optional) Configuration file to override default options
Raw Sequence (FASTQ) files
These are the FASTQ files that need to be mapped to BAM files.
These files are specified in the FASTQ List File.
FASTQ List File
This file specifies the FASTQ files that need to be processed. It maps the FASTQ pairs to the associated Sample ID. Optionally Read Group information for the FASTQ pairs can be specified. If the Read Group information is not specified, it is inferred.
This file is specified either via the command line parameter --list or via the configuration file setting FASTQ_LIST.
The command-line setting takes precedence over the configuration file setting.
The FASTQ list is a tab delimited file that starts with a header line. The columns may be in any order.
Following the header line, there is one line per single-end read and one line per paired-end read (only 1 line per pair).
Required Column Names:
- MERGE_NAME - base name for the resulting BAM file for the sample (used to group multiple fastqs or fastq pairs into a single BAM)
- The SAMPLE column can be specified instead of MERGE_NAME. SAMPLE will be used for both the sample and the base name.
- FASTQ1 - name of the fastq or the first in the pair if paired-end. (Only 1 line per pair)
Optional Column Names:
- FASTQ2 - name of the 2nd fastq in paired-end reads. Specify '.' if the column exists, but this line is single-ended.
- RGID - Read Group ID for this entry
- If this field is not specified, the first line of the fastq will be used to determine the RG.
- If the first line does not match the expected format for determining RG, incrementing numbers per fastq file will be used.
- If this field is not specified, the first line of the fastq will be used to determine the RG.
- SAMPLE - Sample Name for this entry
- If SAMPLE is not specified, MERGE_NAME will be used for the sample name
- LIBRARY - Library for this entry
- If LIBRARY is not specified, the sample name will be used
- CENTER - Center Name for this entry
- If CENTER is not specified, it will default to "unknown"
- PLATFORM - Platform for this entry
- If PLATFORM is not specified, it will default to ILLUMINA
The RGID, SAMPLE, LIBRARY, CENTER, and PLATFORM are used to populate the Read Group information for this entry.
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM Sample1 fastq/S1/F1_R1.fastq.gz fastq/S1/F1_R2.fastq.gz RGID1 SampleID1 Lib1 UM ILLUMINA Sample1 fastq/S1/F2_R1.fastq.gz fastq/S1/F2_R2.fastq.gz RGID1a SampleID1 Lib1 UM ILLUMINA Sample2 fastq/S2/F1_R1.fastq.gz fastq/S2/F1_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA
The --fastq_prefix/FASTQ_PREFIX setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files.
Reference Files
See GotCloud: Genetic Reference and Resource Files for detailed information about the multiple required reference files for the alignment pipeline, including:
- How to obtain default references
- Configuration keys & default values
- How to generate your own references
- How to point GotCloud to your reference files
Required Reference File Types:
Configuration File
The GotCloud configuration file contains the run-time options, including software binaries and command line arguments. A default configuration file is automatically loaded. Users may specify their own configuration file specifying just the values different than the defaults. The configuration file is not required if there are no values to override.
- Default GotCloud configuration file is
gotcloud/bin/gotcloudDefaults.conf - Comments begin with a
# - Format:
KEY = value- where
KEYis the item being set andvalueis its new value
- where
- Some settings can be defined both in the configuration file and on the GotCloud command-line
- command-line options take priority over configuration file settings
- A KEY can be used in another KEY's value by specifying $(KEY)
- Example:
KEY1 = value1KEY2 = $(KEY1)/value2
- When
KEY2is used, it will be equal to:value1/value2
- Example:
Output Directory
- The output directory is required for running GotCloud, so GotCloud knows where to write its output
| Configuration Key | Command-line Flag | Value Description | ||
|---|---|---|---|---|
| OUT_DIR | --outdir | output directory | ||
Reference/Resource Files
- See GotCloud: Genetic Reference and Resource Files for reference/resource file configuration settings
Cluster Configuration
Regardless of the type of cluster system used, GotCloud will wait for each job to complete after launching it.
- For any BATCH_TYPEs that run in batch mode, GotCloud generates a script that will wait until the step is complete before returning
- In a sense, it "fakes" interactive mode for all batch types since it will not proceed until a command is finished
- If you are at UM and are using flux, you can specify either
fluxorpbs
| Configuration Key | Command-line Flag | Value Description | ||
|---|---|---|---|---|
| BATCH_TYPE | --batchtype | type of cluster system | ||
| Valid Values | Command to Launch | Command to Check for Completion | ||
mosix |
mosbatch -E/tmp |
N/A - interactive type | ||
sge |
qsub |
qstat -u $USER
| ||
sgei |
qrsh -now n |
N/A - interactive type | ||
pbs |
qsub |
qstat -u $USER
| ||
slurm |
sbatch |
squeue -u $USER
| ||
slurmi |
|
N/A - interactive type | ||
local |
N/A - local command | N/A - interactive type | ||
| BATCH_OPTS | --batchopts | options to pass to your cluster type, example:
-j36,37,38,39,40,41,45,46,47,48,49 | ||
Recommended Settings
As of GotCloud version 1.16, the alignment pipeline uses bwa mem by default. Prior to version 1.16, the default aligner was bwa aln.
You can override the defaults by setting in your configuration file:
- to use
bwa mem(you do not need to set this in version 1.16 and later since it is the default)
MAP_TYPE = BWA_MEM
- to use
bwa aln(you do not need to set this prior to version 1.16 since it is the default)
MAP_TYPE = BWA
Additional Required Settings
See FASTQ List File for how to set the index file either via command line options or via configuration.
Turning Off Optional Steps
Quality Control steps can be disabled.
To Disable QPLOT, remove qplot from the PER_MERGE_STEPS configuration by setting:
PER_MERGE_STEPS = verifyBamID index recab
To Disable VerifyBamID, remove qplot from the PER_MERGE_STEPS configuration by setting:
PER_MERGE_STEPS = qplot index recab
Optional Configurable Settings
You may want to adjust the amount of memory/threads that are used:
There are additional configurable settings, but these are the ones most likely to be adjusted.
- BWA_THREADS = -t N
- Fill in the N with the number of threads you want BWA to run with, default is 1
- BWA_QUAL = -q N
- Fill in the N with the trim quality you want BWA aln to run with, default is 15. This parameter is only applied to bwa aln. It is not used for BWA_MEM.
- BWA_MEM_OPTS =
- Specify any additional bwa mem options using this parameter.
- SORT_MAX_MEM = 2000000000
- Maximum amount of memory used by samtools sort after running bwa
Running the Alignment Pipeline
Command-Line Options
- help - print usage
- test OUTPUT_DIR - run the test example placing the output in a user specified OUTPUT_DIR. No other options are required.
- outdir OUTPUT_DIR - directory for the output
- May also be specified via OUT_DIR in the configuration file
- Required to be set either via command-line or configuration
- conf CONFIG_FILE - configuration file
- list FASTQ_LIST_FILE_NAME - name of the fastq list file
- May also be specified via FASTQ_LIST in the configuration file
- Required to be set either via command-line or configuration
- ref_dir REFERENCE_DIR - value to set config key REF_DIR to, overriding other values, REF_DIR can then be used inside config files.
- May also be specified via REF_DIR in the configuration file
- ref_prefix REFERENCE_DIR - path to prepend to non-absolute REF paths.
- May also be specified via REF_PREFIX in the configuration file
- fastq_prefix FASTQ_PATH - prefix path to the fastq files specified in the FASTQ_LIST
- May also be specified via FASTQ_PREFIX in the configuration file
- base_prefix BASE_PATH - prefix path to the prepend to fastq/ref files without absolute paths
- May also be specified via BASE_PREFIX in the configuration file
- keepTmp - Do not remove the temporary files (removed by default)
- May also be specified via KEEP_TMP in the configuration file
- keepLog - Do not remove the intermediate log files (removed by default)
- May also be specified via KEEP_LOG in the configuration file
- numjobs N - Replace N with the number of samples that should be processed in parallel
- threads N - Replace N with the number of targets in each makefile that should be run in parallel
- dryrun - Create the Makefile, but do not run it
- maxlocaljobs N - Replace N with the maximum number of jobs that can be run locally (no batchtype specified). Default is 10.
- batchtype TYPE - Tells GotCloud the specified batch type to send jobs to the client nodes
- May also be specified via BATCH_TYPE in the configuration file
- Can be: mosix, slurm, slurmi, pbs, sge, sgei
- batchopts OPTS - Tells GotCloud the options to pass onto the batch system
- May also be specified via BATCH_OPTS in the configuration file
- noPhoneHome - disable the phone home logic
- gotcloudroot DIR - Specifies an alternate path to other gotcloud files rather than using the path to the gotcloud/align.pl.
Note: Command-line options take priority over configuration file settings
Running the Alignment Pipeline
Run gotcloud align with the appropriate command-line parameters.
Example:
gotcloud align --conf config.txt --outdir output
This step generates 1 Makefile per sample in the output/Makefiles/ directory and then automatically runs them. The Makefiles contain all of the information to run each sample.
If you only want to generate the makefiles and not run them, use the --dryrun option. It will generate the Makefiles and print instructions for running the Makefiles.
Each Makefile is independent and can be run in parallel and across a cloud.
On success, you will see:
Processing finished in nn secs with no errors reported
If processing fails part way through, you can pick up where you left off by rerunning gotcloud or the make command.
Alignment Pipeline Output
Upon successful completion of the alignment pipeline, you should see the following files/ subdirectories under the user specified output directory:
- bam.list - file containing sample->BAM mapping that can be used in other GotCloud pipelines
- bams/ - contains the final BAM and bai (BAM index) files
- *.recal.bam
- *.recal.bam.bai
- *.recal.bam.bai.done - temp file indicating this step completed successfully
- *.recal.bam.done - temp file indicating this step completed successfully
- *.recal.bam.metrics - dedup & recalibration log
- *.recal.bam.qemp - recalibration tables
- Makefiles/ - contains the Makefiles and logs used by GotCloud to run the alignment pipeline
- QCFiles/ - contains quality control results if quality control is not disabled
- VerifyBamID Output - see VerifyBamID: A guideline to interpret output files for more information
- *.genoCheck.depthRG - depth distribution of the sequence reads per read group
- *.genoCheck.depthSM - depth distribution of the sequence reads per sample
- *.genoCheck.done - temp file indicating this step completed successfully
- *.genoCheck.selfRG - per-readGroup statistics describing how well each lane matches to the annotated sample
- *.genoCheck.selfSM - main output file containing the contamination estimate; per-sample statistics describing how well the sample matches to the annotated sample
- Check the 'FREEMIX' column for genotype-free estimate of contamination 0-1 scale, the lower, the better
- If [FREEMIX] >= 0.03 and [FREELK1]-[FREELK0] is large, possible contamination
- Qplot Output - see: QPLOT: Diagnose sequencing quality for more info on how to use QPLOT results
- *.qplot.done - temp file indicating this step completed successfully
- *.qplot.R - Rscript that can be used to generate the pdf graphs
- *.qplot.stats - sample statistics
- VerifyBamID Output - see VerifyBamID: A guideline to interpret output files for more information
- tmp/ - contains intermediate files (most are deleted unless --keepTmp is specified)
- *.OK - one OK file per sample; indicates the Sample successfully completed alignment
You should also see a .OK for each Sample in the index file.
If you do not see these .OK files, then your Alignment Pipeline failed.
On success, the bams/ directory contains the final BAMs and bais.