Difference between revisions of "GotCloud: Variant Calling Pipeline"
| Line 141: | Line 141: | ||
</code> | </code> | ||
| − | Replace vc.conf with the | + | * Replace <code>vc.conf</code> with the path/name of the user's configuration file |
| − | + | ** If you are not overriding any defaults, you can alternatively specify <code>--list path/bam.list</code> replacing <code>path/bam.list</code> with the path/name of your BAM list file. | |
| − | If <code>OUT_DIR</code> is not defined in the configuration file, add <code>--outdir</code> followed by the path to the user's desired output directory | + | * Replace <code>2</code> following <code>--numjobs</code> with the number of jobs to be run in parallel |
| − | + | * If <code>OUT_DIR</code> is not defined in the configuration file, add <code>--outdir</code> followed by the path to the user's desired output directory. | |
| − | |||
Revision as of 16:52, 23 October 2014
Back to parent: GotCloud
The Variant Calling Pipeline (previously called 'UMAKE') makes genotype calls from recalibrated BAM files. These genotype calls are output into VCF (Variant Call Format) files.
Running the GotCloud Variant Calling Pipeline
The variant calling pipeline (umake) is run using gotcloud snpcall and gotcloud ldrefine.
Running the Automatic Test
The automatic test runs the variant calling pipeline on a small test set and checks the results against expected results validating that GotCloud is installed correctly.
- Run
snpcallpipeline test:
gotcloud snpcall --test OUTPUT_DIR
- Where OUTPUT_DIR is the directory where you want to store the test results
- If you see
Successfully ran the test case, congratulations!, then you are ready to run snpcall on your own samples.
- Run
ldrefinepipeline test:
gotcloud snpcall --test OUTPUT_DIR
- Where
OUTPUT_DIRis the directory where you want to store the test results - If you see
Successfully ran the test case, congratulations!, then you are ready to run ldrefine on your own samples.
- Where
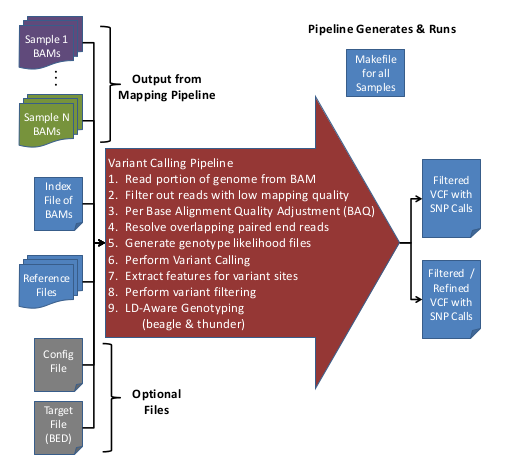
Overview of Variant Calling Pipeline Steps
Here is an overview of the Variant Calling Pipeline:
For more information on the filters applied during the Variant Calling Pipeline, see, GotCloud: Filters.
Input Data
- Aligned/Processed/Recalibrated BAM files
- BAM list file containing Sample IDs & BAM file names
- Reference files
- (Optional) Configuration file to override default options
BAM Files
The BAM files need to be duplicate-marked and base-quality recalibrated in order to obtain high quality SNP calls. Generating these BAM files from original FASTQs is automatically done as part of the Alignment Pipeline of GotCloud.
BAM List File
- Automatically created when running the GotCloud Alignment Pipeline
- Each line of the BAM list file represents a single individual
Columns:
- sample id
- comma separated population labels (optional column)
- BAM File 1 (preferable to have full paths to BAM files)
- BAM File 2 (if more than 1 BAM per sample)
- ...
- # BAM File N (if more than 1 BAM per sample)
[SAMPLE_ID] [COMMA SEPARATED POPULATION LABELS] [BAM_FILE1] [BAM_FILE2] ...
or
[SAMPLE_ID] [BAM_FILE1] [BAM_FILE2] ...
- Notes:
- tab delimited
- multiple BAMs per individual may be provided, but should all be on the same line of the list file
- population label is optional - it will default to
ALL- population is only used by Thunder (part of ldrefine pipeline)
- if all samples are from the same population, population label can be skipped or you can just specify
ALLfor the population label for each sample.
Reference Files
See GotCloud: Genetic Reference and Resource Files for detailed information about the multiple required reference files for the variant calling pipeline, including:
- How to obtain default references
- Configuration keys & default values
- How to generate your own references
- How to point GotCloud to your reference files
Required Reference File Types:
Configuration File
Configuration file contains the run-time options including the software binaries and command line arguments. A default configuration file is automatically loaded. Users must specify their own configuration file specifying just the values different than the defaults.
Comments begin with a #
Format: KEY = value
Where KEY is the item being set and value is its new value
Required User Config Files Settings
The following Config File Settings must be specified by the user:
- CHRS = space separated list of chromosomes you want
- BAM_INDEX = path to the Index File of BAMs
Required on Command-Line or in Config File
The following Command-Line or Config File Settings must be specified by the user:
- --outdir/OUT_DIR= path to desired output directory
Targeted/Exome Sequencing Settings
If you are running Targeted/Exome Sequencing, the user should specify:
- Write loci file when performing pileup
- WRITE_TARGET_LOCI = TRUE
- Specify the output sub-directory to store target information, for example: targetDir
- Should not be a full path as this will co under the OUT_DIR directory.
- TARGET_DIR = targetDir
If all individuals have the same target:
- Specify the single bed file, for example: target.bed
- UNIFORM_TARGET_BED = target.bed
If not all individuals have the same target:
- Specify the file containing the sample id -> bed map, for example: targetMap.txt
- MULTIPLE_TARGET_MAP = targetMap.txt
- Each line of the file contains [SM_ID] [TARGET_BED]
- MULTIPLE_TARGET_MAP = targetMap.txt
Optional Settings:
- Extend the target region by a given number of bases, for example: 50
- OFFSET_OFF_TARGET = 50
Configure Reference Files
See Reference Files for information on how to specify the reference files.
Chromosome X Calling
Making calls on the X chromosome requires the user to specifty a PED file with sex information.
- PED_INDEX = pedfile.ped
Example Configuration File
Example configuration file where reference files happen to be stored in /path/reference, and bam index file in path/freeze5
CHRS = 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 BAM_INDEX = /path/freeze5/freeze5.bam.index ### The BAM index file described above OUT_DIR = /path/freeze5/output ### Directory in which to put all gotcloud output REF = /path/reference/hs37d5.fa ### Reference sequence INDEL_PREFIX = /path/reference/1kg.pilot_release.merged.indels.sites.hg19 ### Known indel sites HM3_VCF = /path/reference/hapmap3_r3_b37.sites.vcf.gz ### HapMap variants (requires tabix index file in same directory) DBSNP_VCF = /path/reference/dbsnp_135.b37.sites.vcf.gz ### dbSNP variants (requires tabix index file in same directory)
Running
Running variant calling is straightforward:
gotcloud snpcall --conf vc.conf --numjobs 2
gotcloud ldrefine --conf vc.conf --numjobs 2
- Replace
vc.confwith the path/name of the user's configuration file- If you are not overriding any defaults, you can alternatively specify
--list path/bam.listreplacingpath/bam.listwith the path/name of your BAM list file.
- If you are not overriding any defaults, you can alternatively specify
- Replace
2following--numjobswith the number of jobs to be run in parallel - If
OUT_DIRis not defined in the configuration file, add--outdirfollowed by the path to the user's desired output directory.
Running on a Cluster
To run on the Cluster, the following settings need to be added to the configuration file:
BATCH_TYPE = batch_type BATCH_OPTS = options to your batch system, as you would normally specify them
Alternatively, --batchtype and --batchopts can be specified on the command line.
Valid values for BATCH_TYPE are: mosix, sge, sgei, slurm, slurmi, pbs, local
- If you are at UM and are using flux, you can specify either
fluxorpbs. sgeiandslurmirun in interactive mode.- For any BATCH_TYPEs that run in batch mode, GotCloud generates a script that will wait until the step is complete before returning.
- In a sense, it "fakes" interactive mode for all batch types since it will not proceed until a command is finished.
Here's the same configuration file we used above but now made to run on a cluster computer with MOSIX.
== Example Configuration File == CHRS = 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 BAM_INDEX = /path/freeze5/freeze5.bam.index OUT_DIR = /path/freeze5/output REF = /path/reference/hs37d5.fa INDEL_PREFIX = /path/reference/1kg.pilot_release.merged.indels.sites.hg19 HM3_VCF = /path/reference/hapmap3_r3_b37.sites.vcf.gz DBSNP_VCF = /path/reference/dbsnp_135.b37.sites.vcf.gz BATCH_TYPE = mosix ### Specify MOSIX as the batch system BATCH_OPTS = -j10,11,12,13 ### Specify available MOSIX compute nodes
Results
If there is a failure, you should see a message like:
make: *** [...] Error 1
Where ... is filled in with other text indicating what step failed.
On SNP Call success, you should see the following output sub-directories under your output directory:
- glfs with a bams & samples subdirectory
- pvcfs with a subdirectory per chromosome and then per region
- split with a subdirectory per chromosome
- vcfs with a subdirectory per chromosome
- (optionally your target directory)
Under the vcf/chrXX directory, there should be:
- chrXX.filtered.sites.vcf
- chrXX.filtered.sites.vcf.norm.log
- chrXX.filtered.sites.vcf.summary
- chrXX.filtered.vcf.gz
- chrXX.filtered.vcf.gz.OK
- chrXX.filtered.vcf.gz.tbi
- chrXX.hardfiltered.sites.vcf
- chrXX.hardfiltered.sites.vcf.log
- chrXX.hardfiltered.sites.vcf.summary
- chrXX.hardfiltered.vcf.gz
- chrXX.hardfiltered.vcf.gz.OK
- chrXX.hardfiltered.vcf.gz.tbi
- chrXX.merged.sites.vcf
- chrXX.merged.stats.vcf
- chrXX.merged.vcf
- chrXX.merged.vcf.OK
The .merged.vcf is the merged together versions of the separate regions in the same chromosome.
The filtered is the merged.vcf after it has been run through filters and is marked with PASS/FAIL.
Under the split/chrXX directory, there should be:
- chrXX.filtered.PASS.split.[N].vcf.gz
- chrXX.filtered.PASS.split.err
- chrXX.filtered.PASS.split.vcflist
- chrXX.filtered.PASS.gz
- subset.OK