Difference between revisions of "LocusZoom Standalone"
| Line 311: | Line 311: | ||
This command creates a database called "my_database.db" and inserts the SNP position table into it. If "my_database.db" had existed already, it would drop the snp_pos table in it, and insert yours in its place. | This command creates a database called "my_database.db" and inserts the SNP position table into it. If "my_database.db" had existed already, it would drop the snp_pos table in it, and insert yours in its place. | ||
| − | One special note about adding SNP position tables: a refsnp_trans table will automatically be created for you, where each SNP maps to itself. If you have a list of SNPs from previous builds that you would like to map to a SNP in the current build, you can then insert your own refsnp_trans table (see | + | One special note about adding SNP position tables: a refsnp_trans table will automatically be created for you, where each SNP maps to itself. If you have a list of SNPs from previous builds that you would like to map to a SNP in the current build, you can then insert your own refsnp_trans table (see below for more information on this table.) |
| + | |||
| + | ==== Inserting refsnp_trans ==== | ||
| + | |||
| + | The refsnp_trans table looks like the following: | ||
| + | |||
| + | {| width="75%" cellspacing="0" cellpadding="5" border="1" class="sortable" | ||
| + | |- | ||
| + | ! scope="col" | rs_orig | ||
| + | ! scope="col" | rs_current | ||
| + | |- | ||
| + | | rs840 || rs715 | ||
| + | |- | ||
| + | | rs1086 || rs940 | ||
| + | |- | ||
| + | | rs1234 || rs1067 | ||
| + | |} | ||
| + | |||
| + | The first column contains SNP names from older genome builds, and the rs_current column contains SNP names from the current genome build (i.e., the build your database file is anchored to.) | ||
| + | |||
| + | Inserting this table into your database is simply then: | ||
| + | |||
| + | <pre> dbmeister.py --db my_database.db --trans my_snp_translations_file </pre> | ||
| + | |||
| + | You will want to execute this command AFTER inserting the snp_pos table, since that command drops the existing translation table. | ||
==== Inserting refFlat ==== | ==== Inserting refFlat ==== | ||
| + | |||
| + | The refFlat table mirrors what is currently supplied by the refFlat table in the UCSC database. The file should look like: | ||
| + | |||
| + | |||
| + | |||
==== Inserting recomb_rate ==== | ==== Inserting recomb_rate ==== | ||
==== Inserting snp_set ==== | ==== Inserting snp_set ==== | ||
| − | + | ||
=== Changing m2zfast.conf settings === | === Changing m2zfast.conf settings === | ||
Revision as of 13:35, 25 May 2010
This page contains information regarding a version of LocusZoom that may be downloaded for personal use. For more information on LocusZoom, see this page.
Requirements
The following software is required:
- Python 2.6 (do not download the 3.0 branch!)
- R 2.10+
- new_fugue, a program for computing LD, written by Goncalo Abecasis.
Currently only Unix/Linux is supported, though Mac OS X should be supported in a future release.
Support for Windows may come at a much later date.
Synopsis
A quick example
First, change directory into examples/. Then, run the following command:
./run_example.py
This script runs the following command for you:
../bin/locuszoom --metal Kathiresan_2009_HDL.txt --refgene FADS1
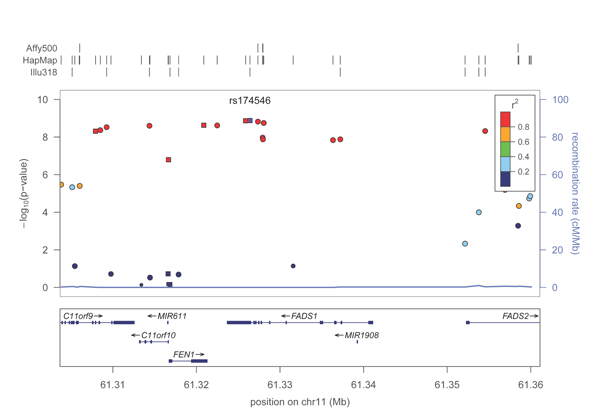
A PDF plot of the FADS1 locus will be created in the directory. It should look roughly like this:
Voila, your first region plot!
Installation
Step 1: Install Python
You will need to install Python on your system if it is not already. Head over to www.python.org to download it. Note that you will want to make sure to download the latest from the 2.x branch, and not the 3.0 one.
Step 2: Install R
R is also required for generating the plots. You can download R at www.r-project.org. Version 2.10 or greater is required.
Step 3: Install new_fugue
New_fugue is a program that calculates linkage disequilibrium measures from genotype files. While installing new_fugue is optional, we highly recommend it as it makes the process of generating plots much easier. If you opt to skip installing new_fugue, you will need to provide your own computed LD files for each region that you want to plot.
New_fugue can be downloaded from here.
Once downloaded, extract the tar file using:
tar zxf /path/to/new_fugue.tar.gz
Change into the generic-new_fugue directory that is created, and run:
make install
Step 4: Install LocusZoom
LocusZoom is provided as a tar archive which contains the following:
- the LocusZoom python application
- the R script used for generating plots
- genotype files (used for computing LD) from hapmap and 1000G (build hg18 only)
- a SQLite database file containing tables describing SNP positions, SNP annotations, gene and exon locations, and recombination rates (build hg18 only)
Simply unpack the tar to your directory of choice by doing the following:
cd <directory where you want to place locuszoom> tar zxf /path/to/locuszoom.tgz
The tar archive will extract into the following directory structure:
- locuszoom/
- bin/
- locuszoom (this is the locuszoom "executable")
- locuszoom.R (the R script which is used by locuszoom for creating the plots)
- conf/ (configuration file located here)
- data/
- database/ (SQLite file located here)
- hapmap/ (hapmap genotype files)
- 1000G/ (1000G genotype files)
- src/ (source code for locuszoom)
- bin/
It is important that this directory structure remain intact. To make launching locusoom easier, you could create a link to it from /usr/local/bin, for example:
ln -s bin/locuszoom /usr/local/bin/locuszoom
Input
Association results file ("metal" file)
The main input to LocusZoom is a file containing results from an association scan or meta-analysis. The file must have 2 things: markers (SNPs), and p-values. The file should look something like this:
| MarkerName | P-value |
|---|---|
| rs1 | 0.423 |
| rs2 | 1.23e-04 |
| rs3 | 9.4e-390 |
The file should be tab-delimited, though this can be changed using the --delim option.
This file should be passed to locuszoom using the --metal option.
If your marker and p-value column names are not "MarkerName" and "P-value", you may set them with --markercol and --pvalcol options.
P-values of any magnitude are supported in scientific notation (we use an arbitrary precision library built-in to python, and transform p-values to the log scale.) If you've already transformed your p-values to the log scale, simply use --no-transform and LocusZoom will not transform them.
Region
You can specify the region to plot in any one of the following ways:
- A reference SNP and flanking region
--refsnp <your snp> --flank 500kb
- A reference SNP and chromosome/start/stop specification
--refsnp <your snp> --chr # --start <base position> --end <base position>
- A gene and flanking region
--refgene <your gene> --flank 250kb
The flank is computed as +/- from the transcription start/end of the gene. From this region, LocusZoom will find the SNP with the most significant p-value, and use this as the reference SNP.
- A gene and chromosome/start/stop specification
--refgene <your gene> --chr # --start <base position> --end <base position>
This method is similar to the above, except that an exact region is specified. The SNP with the most significant p-value in this region will be used.
- A chromosome/start/stop specification
--chr # --start <base position> --end <base position>
Once again, the SNP with the most significant p-value will be used in this region.
Batch mode
LocusZoom provides two fast methods for generating plots for a large number of regions:
--hits, which parses a file for SNP names (rs#) and creates a plot for each one.--hitspec, which reads a batch mode specification file.
To use --hits, you need only provide any text file that has SNP names (of the rs### variety) present in the file. LocusZoom will extract SNPs from this file, regardless of formatting. Note that this file shouldn't be very large, or the parsing procedure could take a long time.
For a more thorough specification of each plot you would like to create, use --hitspec. For this option, you must provide a text file of the following format:
| Column | Description |
|---|---|
| snp | Can be either a SNP, or gene. |
| chr | Chromosome |
| start | Start position to plot. |
| stop | Stop position. |
| flank | Flank for region. Can be given instead of chr/start/stop. |
| run | Should this row be read? Should be "yes" or "no". |
| m2zargs | List of arguments for customizing plots. You can find a list of them here: Commonly Used LocusZoom Options |
The file should be delimited by whitespace (tab, space, multiple spaces), and the header must exist, with column names exactly as specified in the table above. As an example, consider the following file:
| snp | chr | start | stop | flank | run | m2zargs |
|---|---|---|---|---|---|---|
| rs7983146 | NA | NA | NA | 500kb | yes | title="My favorite SNP" |
| TCF7L2 | NA | NA | NA | 1.25MB | yes | title="TCF7L2 Region" showRecomb=F |
| rs7957197 | 12 | 119503590 | 120322280 | NA | yes | showAnnot=F |
The first row would plot rs7983146 as the reference SNP, and a region of 500kb on either side of it. The plot title would read "My favorite SNP."
The second row would plot 1.25 MB on either side of TCF7L2's transcription start and stop. The SNP with the most significant p-value in your --metal file will be used as the reference SNP. The plot title would read "TCF7L2 Region", and the recombination overlay would be disabled using showRecomb=F.
The third row would plot rs7957197 as the reference SNP, but here we've specifically designated the region to plot, which is chr12:119503590-120322280. We've also disabled showing SNP annotations with showAnnot=F.
User-supplied LD
LocusZoom by default automatically computes LD between the reference SNP and all other SNPs within each region to be plotted. However, in some instances, you may wish to provide your own file with LD information. This can be done with the --ld option, which requires a file of the following format:
| Column | Description |
|---|---|
| snp1 | Any SNP in your plotting region. |
| snp2 | Should always be the reference SNP in the region. |
| dprime | D' between snp2 (reference SNP) and snp1. |
| rsquare | r2 between snp2 (reference SNP) and snp1. |
The file should be whitespace delimited, and the header (column names shown above) must exist.
Output
LocusZoom will produce a directory for each plot that contains the plot itself, along with a number of temporary files containing information on your particular region. The plot will be a PDF, named with the chr#:start-stop that was plotted.
If you only want the PDF itself, and don't want the other files, you can use the --plotonly option.
Each directory (or PDF, in the case of --plotonly) will have the date included to avoid collisions with previous plots - this behavior can be disabled using --no-date.
You can further customize the directory/PDF names that are created by using the --prefix <name> option. This will append a text string at the beginning of each directory/PDF that is created.
Options
LocusZoom has a number of command line options, described in the table below.
| Option | Description |
|---|---|
| Important settings | |
| --metal | This is the data file to provide. Files generated by the meta-analysis program METAL are already formatted appropriately. If your data is not from METAL, it is very simple to format it (see #Input.) |
| --delim | Delimiter for the data file. This defaults to tab, but can be anything. For ease of specification, you can use the following shortcuts: --delim tab, --delim space, --delim comma. |
| --pvalcol | Name of p-value column in the --metal file. |
| --markercol | Name of the SNP column in the --metal file. |
| --refsnp | Reference SNP to be used in the plot. |
| --refgene | Specify a gene instead of a reference SNP. This will plot a region near a gene, and automatically find the SNP with the most significant p-value to use as the reference SNP. |
| --flank | Specify the region near a reference SNP or gene as a "flank", instead of having to specify chr/start/stop explicitly. This can be specified in bases, kilobases, or megabases. Examples: 500kb, 1MB, 100141 |
| --chr, --start, --stop | Specify chromosome/start/stop as the exact interval to plot. If no --refsnp is specified, the SNP with the most significant p-value in the region will be used as the reference SNP. |
| Optional settings | |
| --build | Human genome build. This defaults to "hg18", and is the only build we provide data for currently. You can supply your own build-specific data by modifying the conf file, and creating your own SQLite database (see *LINK HERE*). |
| --ld | Provide a file specifying LD between your reference SNP and all SNPs within the region you wish to plot. You only need to supply this file if you have created LD specifically for your purposes (perhaps a different population or genome build.) Otherwise, LD is computed automatically for you. |
| --source | Source to use for genotypes when using LD. Currently, we support "1000G" and "hapmap". |
| --pop | Population to use when computing LD. Currently, when we support "CEU" for 1000G, and "CEU", "YRI", and "JPT+CHB" for hapmap. |
| --snpset | Rug of SNPs to create at the top of the plot. Defaults to the Illumina 1M chip currently. |
| --plotonly | Create only a PDF of the plot, and remove all temporary files/directories created during plotting. |
| --no-transform | LocusZoom supports arbitrary precision p-values. However, if your p-values have already been transformed to the log scale, you can use this option to stop LocusZoom from automatically transforming them. |
| --prefix | Places a text string at the beginning of each plot or directory created. This is mainly used to denote different batches of plots - for example, you could use --prefix using_ceu to denote these plots are computed using CEU LD information. |
| --db | SQLite database file to use. This is set in the conf file by default, but can be changed on the command line if desired. |
Advanced configuration
Creating a SQLite database
As a starting point, we provide a SQLite database with the following tables:
- snp_pos: SNP positions
- refFlat: gene information (exons, transcription start/stops, etc.)
- recomb_rate: recombination rates from hapmap phase 2
- snp_set: maps each SNP to a "set" - for example, all SNPs on the Illumina 1M chip
- refsnp_trans: a table that maps SNPs from previous builds to the current build
This database is anchored to the UCSC hg18 build of the human genome.
To create your own database, we provide a script bin/dbmeister.py that can insert these tables for you. We recommend creating your own database file, rather than inserting tables into the default LocusZoom database. This script is capable of using python's built-in sqlite support, but for faster insertion of tables (about 2x faster), we recommend installing sqlite3 from www.sqlite.org.
Inserting snp_pos
First, create a file that looks like the following:
| snp | chr | pos |
|---|---|---|
| rs38343 | 1 | 93919141 |
| rs918141 | 7 | 763263 |
| chr4:9181 | 4 | 9181 |
The file should be: tab-delimited, must have a header, and the columns should be exactly in that order.
Now, you can create your own database, and insert this file by using:
dbmeister.py --db my_database.db --snp_pos my_snp_pos_file
This command creates a database called "my_database.db" and inserts the SNP position table into it. If "my_database.db" had existed already, it would drop the snp_pos table in it, and insert yours in its place.
One special note about adding SNP position tables: a refsnp_trans table will automatically be created for you, where each SNP maps to itself. If you have a list of SNPs from previous builds that you would like to map to a SNP in the current build, you can then insert your own refsnp_trans table (see below for more information on this table.)
Inserting refsnp_trans
The refsnp_trans table looks like the following:
| rs_orig | rs_current |
|---|---|
| rs840 | rs715 |
| rs1086 | rs940 |
| rs1234 | rs1067 |
The first column contains SNP names from older genome builds, and the rs_current column contains SNP names from the current genome build (i.e., the build your database file is anchored to.)
Inserting this table into your database is simply then:
dbmeister.py --db my_database.db --trans my_snp_translations_file
You will want to execute this command AFTER inserting the snp_pos table, since that command drops the existing translation table.
Inserting refFlat
The refFlat table mirrors what is currently supplied by the refFlat table in the UCSC database. The file should look like: