Difference between revisions of "LocusZoom Standalone"
| (42 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
This page contains information regarding a version of LocusZoom that may be downloaded for personal use. For more information on LocusZoom, see this [[LocusZoom|page]]. | This page contains information regarding a version of LocusZoom that may be downloaded for personal use. For more information on LocusZoom, see this [[LocusZoom|page]]. | ||
| + | |||
| + | To be notified of future changes to LocusZoom, you can join our [http://groups.google.com/group/locuszoom Google Group]. | ||
== Quick Start (Requirements) == | == Quick Start (Requirements) == | ||
| Line 12: | Line 14: | ||
The following software is required: | The following software is required: | ||
| − | *[http://www.python.org/download/ Python 2. | + | *[http://www.python.org/download/ Python 2.7+] (do '''not''' download the 3.0 branch!) |
| − | *[http://www.r-project.org/ R | + | *[http://www.r-project.org/ R 3.0+]. Note that if using R 3.1, you must install LocusZoom 1.3 (previous versions will fail.) |
| + | |||
| + | |||
| + | The following software is optional but recommended: | ||
| + | |||
*[[New Fugue|new_fugue]], a program for computing LD, written by Goncalo Abecasis. | *[[New Fugue|new_fugue]], a program for computing LD, written by Goncalo Abecasis. | ||
| + | *[http://pngu.mgh.harvard.edu/~purcell/plink/ PLINK] | ||
| + | *[http://samtools.sourceforge.net/ tabix], downloaded with samtools | ||
| + | |||
| + | |||
| + | The following R packages are optional but recommended: | ||
| + | *[http://cran.r-project.org/web/packages/gridExtra/index.html gridExtra] (used for creating summary tables of GWAS hits / fine-mapping SNPs as additional pages in the PDF) | ||
| − | |||
| − | + | For the latest stable LocusZoom package, see our [https://github.com/statgen/locuszoom-standalone download] page. The current version is '''1.3''', released on June 20th, 2014. | |
| − | Support for Windows may come at a much later date. | + | Currently only '''Unix/Linux''' is supported, though Mac OS X should be supported in a future release. Support for Windows may come at a much later date. |
== Synopsis == | == Synopsis == | ||
| Line 30: | Line 41: | ||
A PDF plot of the FADS1 locus will be created in the directory. It should look roughly like this: | A PDF plot of the FADS1 locus will be created in the directory. It should look roughly like this: | ||
| − | [[Image:FADS1 small.png]] | + | [[Image:FADS1 small.png]] |
| − | |||
| − | |||
== Download == | == Download == | ||
| − | See our [https://statgen | + | See our [https://github.com/statgen/locuszoom-standalone download] page for links to the latest as well as previous releases. |
== Installation == | == Installation == | ||
| Line 42: | Line 51: | ||
=== Step 1: Install Python === | === Step 1: Install Python === | ||
| − | You will need to install Python on your system if it is not already. Head over to [http://www.python.org www.python.org] to download it. Note that you will want to make sure to download the latest from the 2.x branch, and '''not''' the 3. | + | You will need to install Python on your system if it is not already. Head over to [http://www.python.org www.python.org] to download it. Note that you will want to make sure to download the latest from the 2.x branch, and <span style="color:#ff0000">'''*not*'''</span> the 3.x one. |
=== Step 2: Install R === | === Step 2: Install R === | ||
| Line 48: | Line 57: | ||
R is also required for generating the plots. You can download R at [http://www.r-project.org/ www.r-project.org]. Version 2.10 or greater is required. | R is also required for generating the plots. You can download R at [http://www.r-project.org/ www.r-project.org]. Version 2.10 or greater is required. | ||
| − | === Step 3: Install new_fugue | + | === Step 3: Install LD calculation software (optional) === |
| + | |||
| + | * If you wish to calculate from hg18 sources (hapmap, earlier releases of 1000G): install '''new_fugue''' (see below.) | ||
| + | * If you wish to calculate from hg19 sources (latest 1000G): install '''PLINK''' (see below.) | ||
| + | * If you plan to supply your own LD files per region, or calculate LD directly from VCF files: install nothing! See options for --ld and --ld-vcf. | ||
| + | |||
| + | ==== new_fugue ==== | ||
New_fugue is a program that calculates linkage disequilibrium measures from genotype files. While installing new_fugue is optional, we highly recommend it as it makes the process of generating plots much easier. If you opt to skip installing new_fugue, you will need to provide your own computed LD files for each region that you want to plot. | New_fugue is a program that calculates linkage disequilibrium measures from genotype files. While installing new_fugue is optional, we highly recommend it as it makes the process of generating plots much easier. If you opt to skip installing new_fugue, you will need to provide your own computed LD files for each region that you want to plot. | ||
| Line 61: | Line 76: | ||
You may need administrator rights to install this program. | You may need administrator rights to install this program. | ||
| − | === Step | + | ==== PLINK ==== |
| + | |||
| + | PLINK is now used to calculate LD for all future LD sources / populations that we may add. The program new_fugue (above) is used to calculate LD from older sources (such as hapmap) and older builds (such as hg18) where LD files are sufficiently small. | ||
| + | |||
| + | You can download PLINK and find instructions for installing it [http://pngu.mgh.harvard.edu/~purcell/plink/download.shtml here]. | ||
| + | |||
| + | === Step 5: Install tabix === | ||
| + | |||
| + | Tabix is used to quickly extract regions from bgzipped and tabix-indexed files. It is used in LocusZoom to extract regions from VCF files when calculating LD, and for extracting from EPACTS result files. | ||
| + | |||
| + | It can be downloaded from the sourceforge site [http://samtools.sourceforge.net/ here] or directly to the download site [http://sourceforge.net/projects/samtools/files/ here]. | ||
| + | |||
| + | === Step 6: Install LocusZoom === | ||
LocusZoom is provided as a tar archive which contains the following: | LocusZoom is provided as a tar archive which contains the following: | ||
| − | * | + | *The LocusZoom python application |
| − | * | + | *The R script used for generating plots |
| − | *Human genome '''build hg18''' data, including: | + | *Human genome '''build hg18 and hg19''' data, including: |
**genotype files (used for computing LD) from HapMap and 1000G | **genotype files (used for computing LD) from HapMap and 1000G | ||
**a SQLite database file containing tables describing SNP positions, SNP annotations, gene and exon locations, and recombination rates | **a SQLite database file containing tables describing SNP positions, SNP annotations, gene and exon locations, and recombination rates | ||
| Line 81: | Line 108: | ||
***locuszoom (this is the locuszoom "executable") | ***locuszoom (this is the locuszoom "executable") | ||
***locuszoom.R (the R script which is used by locuszoom for creating the plots) | ***locuszoom.R (the R script which is used by locuszoom for creating the plots) | ||
| + | ***dbmeister.py (script for creating custom user databases) | ||
| + | ***lzupdate.py (script for creating an updated copy of the provided locuszoom database) | ||
**conf/ (configuration file located here) | **conf/ (configuration file located here) | ||
**data/ | **data/ | ||
| Line 97: | Line 126: | ||
For computing LD: | For computing LD: | ||
*[ftp://ftp.hapmap.org/hapmap/genotypes/2008-10_phaseII/ HapMap genotypes] for populations CEU, YRI, and JPT+CHB. | *[ftp://ftp.hapmap.org/hapmap/genotypes/2008-10_phaseII/ HapMap genotypes] for populations CEU, YRI, and JPT+CHB. | ||
| − | *[ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/release/2010_03/pilot1 1000 Genomes genotypes] | + | *[ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/release/2010_03/pilot1 1000 Genomes genotypes] |
For SNP, gene, and exon positions: | For SNP, gene, and exon positions: | ||
| Line 104: | Line 133: | ||
For annotation: | For annotation: | ||
| − | *We | + | *We use various sources including RefSeq Genes (refFlat), TFBS Conserved (tfbsConsSites), and Conservation (phaseConsElements44wayPlacental), all available from the [http://genome.usc.edu UCSC Genome Browser]. |
*[ftp://ftp.hapmap.org/hapmap/recombination/2008-03_rel22_B36/rates/ Recombination rates from HapMap]. | *[ftp://ftp.hapmap.org/hapmap/recombination/2008-03_rel22_B36/rates/ Recombination rates from HapMap]. | ||
| + | |||
| + | For GWAS hits: | ||
| + | *We use the NHGRI GWAS catalog, available at [http://www.genome.gov/gwastudies/ genome.gov] | ||
== Input == | == Input == | ||
| − | === Association results file | + | === Association results file === |
| + | |||
| + | LocusZoom requires an association results file similar in formatting to what METAL or EPACTS provides. | ||
| + | |||
| + | ==== METAL formatted file ==== | ||
| − | + | The file must have 2 columns: markers (SNPs), and p-values. The file should look something like this: | |
<br> | <br> | ||
| Line 126: | Line 162: | ||
| align="center" | 1.23e-04 | | align="center" | 1.23e-04 | ||
|- | |- | ||
| − | | align="center" | | + | | align="center" | chr4:401141 |
| align="center" | 9.4e-390 | | align="center" | 9.4e-390 | ||
|} | |} | ||
<br> | <br> | ||
| + | |||
| + | Markers can be either rsIDs or chr:pos format (see above). | ||
This file should be passed to locuszoom using the <code>--metal</code> option. | This file should be passed to locuszoom using the <code>--metal</code> option. | ||
| Line 139: | Line 177: | ||
P-values of any magnitude are supported in scientific notation (we use an arbitrary precision library built-in to python, and transform p-values to the log scale.) If you've already transformed your p-values to the log scale, simply use <code>--no-transform</code> and LocusZoom will not transform them. | P-values of any magnitude are supported in scientific notation (we use an arbitrary precision library built-in to python, and transform p-values to the log scale.) If you've already transformed your p-values to the log scale, simply use <code>--no-transform</code> and LocusZoom will not transform them. | ||
| + | |||
| + | ==== EPACTS formatted file ==== | ||
| + | |||
| + | The file can come directly from [[EPACTS]], or simply be formatted similarly to the following: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | ! scope="col" | #CHROM | ||
| + | ! scope="col" | BEGIN | ||
| + | ! scope="col" | END | ||
| + | ! scope="col" | MARKER_ID | ||
| + | ! scope="col" | NS | ||
| + | ! scope="col" | AC | ||
| + | ! scope="col" | CALLRATE | ||
| + | ! scope="col" | MAF | ||
| + | ! scope="col" | PVALUE | ||
| + | ! scope="col" | SCORE | ||
| + | ! scope="col" | N.CASE | ||
| + | ! scope="col" | N.CTRL | ||
| + | ! scope="col" | AF.CASE | ||
| + | ! scope="col" | AF.CTRL | ||
| + | |- | ||
| + | | 1 || 15903 || 15903 || 1:15903_G/GC || 2657 || 3892.2 || 1 || 0.26757 || 0.36771 || 0.90077 || 1326 || 1331 || 1.4688 || 1.4609 | ||
| + | |- | ||
| + | | 1 || 19190 || 19191 || 1:19190_GC/G || 2657 || 823.65 || 1 || 0.155 || 0.67173 || 0.42378 || 1326 || 1331 || 0.3115 || 0.30849 | ||
| + | |- | ||
| + | | 1 || 20316 || 20317 || 1:20316_GA/G || 2657 || 1005.3 || 1 || 0.18917 || 0.50804 || 0.66189 || 1326 || 1331 || 0.38062 || 0.37607 | ||
| + | |- | ||
| + | | 1 || 30967 || 30970 || 1:30967_CCCA/C || 2657 || 435.35 || 1 || 0.081925 || 0.08848 || -1.7035 || 1326 || 1331 || 0.16007 || 0.16762 | ||
| + | |- | ||
| + | | 1 || 51972 || 51975 || 1:51972_GGAC/G || 2657 || 207.8 || 1 || 0.039104 || 0.51638 || -0.64893 || 1326 || 1331 || 0.077187 || 0.079226 | ||
| + | |- | ||
| + | | 1 || 53138 || 53140 || 1:53138_TAA/T || 2657 || 216.2 || 1 || 0.040685 || 0.55679 || 0.58762 || 1326 || 1331 || 0.083145 || 0.079602 | ||
| + | |- | ||
| + | | 1 || 54421 || 54421 || 1:54421_A/G || 2657 || 179.45 || 1 || 0.033769 || 0.73592 || 0.33726 || 1326 || 1331 || 0.068213 || 0.066867 | ||
| + | |- | ||
| + | | 1 || 66221 || 66221 || 1:66221_A/AT || 2657 || 664.45 || 1 || 0.12504 || 0.48676 || 0.69547 || 1326 || 1331 || 0.25366 || 0.24651 | ||
| + | |- | ||
| + | | 1 || 66222 || 66223 || 1:66222_TA/T || 2657 || 470.3 || 1 || 0.088502 || 0.64258 || 0.4641 || 1326 || 1331 || 0.17941 || 0.17461 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | The chrom, start, end, marker ID, and p-value columns must all be present. The file must be tab-delimited. | ||
| + | |||
| + | To load this file, use --epacts. | ||
| + | |||
| + | <span style="color:#00CC33">'''Note'''</span>: LocusZoom (as of 1.3) will now use the tabix index for the EPACTS file if it exists and if tabix is intalled on your system. This results in much faster loading of EPACTS files and should absolutely be used if possible. | ||
| + | |||
| + | <span style="color:#ff0000">'''Warning'''</span>: The "test" version of EPACTS changed the format of the output. To make LZ work, you'll also need to add <code>--epacts-beg-col BEG</code> to your command line. | ||
| + | |||
| + | ==== Reading from STDIN ==== | ||
| + | |||
| + | If you have a quick way of pulling out regions from your association results to plot (such as with tabix), you can pass the data directly to locuszoom on STDIN by specifying the file as "-". For example: | ||
| + | |||
| + | <pre> | ||
| + | tabix -h my_file.gz 1:1-10000 | locuszoom --metal - --refgene TCF7L2 | ||
| + | </pre> | ||
=== Region === | === Region === | ||
| Line 146: | Line 241: | ||
*A reference SNP and flanking region | *A reference SNP and flanking region | ||
<pre> --refsnp <your snp> --flank 500kb </pre> | <pre> --refsnp <your snp> --flank 500kb </pre> | ||
| + | |||
*A reference SNP and chromosome/start/stop specification | *A reference SNP and chromosome/start/stop specification | ||
<pre> --refsnp <your snp> --chr # --start <base position> --end <base position> </pre> | <pre> --refsnp <your snp> --chr # --start <base position> --end <base position> </pre> | ||
| + | |||
| + | *A reference SNP and gene: | ||
| + | <pre> --refsnp <your snp> --refgene <your gene> </pre> | ||
| + | |||
| + | This will use the reference gene as the plotting boundaries. You can extend the boundaries by also including --flank. | ||
| + | |||
*A gene and flanking region | *A gene and flanking region | ||
<pre> --refgene <your gene> --flank 250kb </pre> | <pre> --refgene <your gene> --flank 250kb </pre> | ||
| Line 164: | Line 266: | ||
We supply genotype files for computing LD between the reference SNP and all other SNPs within the region you are plotting. The tables below show the supported combinations of LD source, population, and build. Note that you can always provide your own LD files, see [[#User-supplied_LD|User-supplied LD]] for more information. | We supply genotype files for computing LD between the reference SNP and all other SNPs within the region you are plotting. The tables below show the supported combinations of LD source, population, and build. Note that you can always provide your own LD files, see [[#User-supplied_LD|User-supplied LD]] for more information. | ||
| − | '''1000G''' | + | '''1000G''' |
{| width="75%" cellspacing="0" cellpadding="5" border="1" | {| width="75%" cellspacing="0" cellpadding="5" border="1" | ||
|- | |- | ||
| − | ! | + | ! align="left" scope="col" | Release |
| − | ! | + | ! align="left" scope="col" | Build |
| − | ! | + | ! align="left" scope="col" | Population |
| − | ! | + | ! align="left" scope="col" | LocusZoom Arguments |
| + | |- | ||
| + | | March 2012 | ||
| + | | hg19 | ||
| + | | ASN | ||
| + | | --pop ASN --build hg19 --source 1000G_March2012 | ||
| + | |- | ||
| + | | March 2012 | ||
| + | | hg19 | ||
| + | | AFR | ||
| + | | --pop AFR--build hg19 --source 1000G_March2012 | ||
| + | |- | ||
| + | | March 2012 | ||
| + | | hg19 | ||
| + | | EUR | ||
| + | | --pop EUR --build hg19 --source 1000G_March2012 | ||
| + | |- | ||
| + | | March 2012 | ||
| + | | hg19 | ||
| + | | AMR | ||
| + | | --pop AMR --build hg19 --source 1000G_March2012 | ||
| + | |- | ||
| + | | Nov 2010 | ||
| + | | hg19 | ||
| + | | ASN | ||
| + | | --pop ASN --build hg19 --source 1000G_Nov2010 | ||
| + | |- | ||
| + | | Nov 2010 | ||
| + | | hg19 | ||
| + | | AFR | ||
| + | | --pop AFR --build hg19 --source 1000G_Nov2010 | ||
|- | |- | ||
| − | | | + | | Nov 2010 |
| + | | hg19 | ||
| + | | EUR | ||
| + | | --pop EUR --build hg19 --source 1000G_Nov2010 | ||
|- | |- | ||
| − | | June 2010 | + | | June 2010 |
| + | | hg18 | ||
| + | | CEU | ||
| + | | --pop CEU --build hg18 --source 1000G_June2010 | ||
|- | |- | ||
| − | | June 2010 | + | | June 2010 |
| + | | hg18 | ||
| + | | YRI | ||
| + | | --pop YRI --build hg18 --source 1000G_June2010 | ||
|- | |- | ||
| − | | | + | | June 2010 |
| + | | hg18 | ||
| + | | JPT+CHB | ||
| + | | --pop JPT+CHB --build hg18 --source 1000G_June2010 | ||
|- | |- | ||
| − | | August 2009 | + | | August 2009 |
| + | | hg18 | ||
| + | | CEU | ||
| + | | --pop CEU --build hg18 --source 1000G_Aug2009 | ||
|- | |- | ||
| − | | August 2009 || hg18 || JPT+CHB | + | | August 2009 |
| + | | hg18 | ||
| + | | YRI | ||
| + | | --pop YRI --build hg18 --source 1000G_Aug2009 | ||
| + | |- | ||
| + | | August 2009 | ||
| + | | hg18 | ||
| + | | JPT+CHB | ||
| + | | --pop JPT+CHB --build hg18 --source 1000G_Aug2009 | ||
|} | |} | ||
| − | + | <br> '''HapMap Phase II''' | |
| − | '''HapMap Phase II''' | ||
{| width="75%" cellspacing="0" cellpadding="5" border="1" | {| width="75%" cellspacing="0" cellpadding="5" border="1" | ||
|- | |- | ||
| − | ! | + | ! align="left" scope="col" | Build |
| − | ! | + | ! align="left" scope="col" | Population |
| − | ! | + | ! align="left" scope="col" | LocusZoom Arguments |
|- | |- | ||
| − | | hg18 | + | | hg18 |
| + | | CEU | ||
| + | | --pop CEU --build hg18 --source hapmap | ||
|- | |- | ||
| − | | hg18 | + | | hg18 |
| + | | YRI | ||
| + | | --pop YRI--build hg18 --source hapmap | ||
|- | |- | ||
| − | | hg18 | + | | hg18 |
| + | | JPT+CHB | ||
| + | | --pop JPT+CHB --build hg18 --source hapmap | ||
|} | |} | ||
| + | |||
=== Batch mode === | === Batch mode === | ||
| − | LocusZoom provides | + | LocusZoom provides a batch mode for generating plots for a large number of regions: |
| − | |||
*<code>--hitspec </code>, which reads a batch mode specification file. | *<code>--hitspec </code>, which reads a batch mode specification file. | ||
| − | + | For this option, you must provide a text file of the following format: | |
| − | |||
| − | |||
{| width="75%" cellspacing="0" cellpadding="5" border="1" | {| width="75%" cellspacing="0" cellpadding="5" border="1" | ||
| Line 225: | Line 383: | ||
|- | |- | ||
| start | | start | ||
| − | | Start position to plot. | + | | Start position to display on plot. |
|- | |- | ||
| − | | | + | | end |
| − | | | + | | End position to display on plot. |
|- | |- | ||
| flank | | flank | ||
| Line 285: | Line 443: | ||
The second row would plot 1.25 MB on either side of TCF7L2's transcription start and stop. The SNP with the most significant p-value in your <code>--metal</code> file will be used as the reference SNP. The plot title would read "TCF7L2 Region", and the recombination overlay would be disabled using showRecomb=F. | The second row would plot 1.25 MB on either side of TCF7L2's transcription start and stop. The SNP with the most significant p-value in your <code>--metal</code> file will be used as the reference SNP. The plot title would read "TCF7L2 Region", and the recombination overlay would be disabled using showRecomb=F. | ||
| − | The third row would plot rs7957197 as the reference SNP, but here we've specifically designated the region to plot, which is chr12:119503590-120322280. We've also disabled showing SNP annotations with showAnnot=F. | + | The third row would plot rs7957197 as the reference SNP, but here we've specifically designated the region to plot, which is chr12:119503590-120322280. We've also disabled showing SNP annotations with showAnnot=F. |
=== User-supplied LD === | === User-supplied LD === | ||
| Line 312: | Line 470: | ||
<br> | <br> | ||
| + | |||
| + | The dprime column can be all missing if it is not known. Rsquare must be present, and must be valid data. | ||
The file should be whitespace delimited, and the header (column names shown above) must exist. | The file should be whitespace delimited, and the header (column names shown above) must exist. | ||
| + | |||
| + | === Supply VCF files for calculating LD === | ||
| + | |||
| + | You can give LocusZoom a VCF file directly to use for calculating LD: | ||
| + | |||
| + | <pre> | ||
| + | locuszoom --ld-vcf my_genotypes.vcf.gz ... | ||
| + | </pre> | ||
| + | |||
| + | This option takes the place of having to supply per-region pre-calculated LD (--ld) or having to specify --pop and --source for calculating LD from genotype files supplied by LZ. | ||
| + | |||
| + | <span style="color:#FF6600">'''Warning: '''</span> The VCF file must also have a [http://samtools.sourceforge.net/tabix.shtml tabix] index located in the same directory. For the above example, the tabix index "my_genotypes.vcf.gz.tbi" must exist. | ||
| + | |||
| + | |||
| + | You can also calculate D' from phased VCF files: | ||
| + | |||
| + | <pre> | ||
| + | locuszoom --ld-vcf my_genotypes.vcf.gz --ld-measure dprime ... | ||
| + | </pre> | ||
| + | |||
| + | The default measure is "rsquared". | ||
| + | |||
| + | In version 1.3, if you have VCF files separated out by chromosome, you can create a JSON file mapping chromosome name --> VCF file, and provide the JSON file to --ld-vcf. For example, the JSON file could look like: | ||
| + | |||
| + | <pre> | ||
| + | { | ||
| + | "X": "/path/to/X.vcf.gz", | ||
| + | "Y": "/path/to/Y.vcf.gz", | ||
| + | "MT": "/path/to/MT.vcf.gz", | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | And then pass it directly using <code>locuszoom --ld-vcf my_vcfs.json</code>. | ||
| + | |||
| + | == Optional Input == | ||
| + | |||
| + | === Plotting LD with additional reference SNPs === | ||
| + | |||
| + | LocusZoom can now show LD with multiple SNPs in a region (for example, you might want to show LD with a number of SNPs from a conditional analysis.) | ||
| + | |||
| + | You give LocusZoom the usual reference SNP (used for centering the plot and calculating the region) but an additional set of lead/reference SNPs as well. | ||
| + | |||
| + | For all other SNPs not in the "lead SNP set" of { reference SNP, additional reference SNPs }, LZ will find which of the lead SNPs it is in highest LD with, and color it to match that lead SNP. The extent of LD with the lead SNP is shown by a gradient of color. | ||
| + | |||
| + | |||
| + | As an example: | ||
| + | |||
| + | <syntaxhighlight lang="bash"> | ||
| + | locuszoom --metal <DIAGRAM T2D results> --refsnp "rs231362" --add-refsnps "rs163184" | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | |||
| + | Will generate the following plot: | ||
| + | |||
| + | [[File:New lz cond only.png|700px]] | ||
| + | |||
| + | |||
| + | The following options are available for changing the style of these types of plots: | ||
| + | |||
| + | {| width="85%" cellspacing="0" cellpadding="5" border="1" | ||
| + | |- | ||
| + | ! scope="col" | Option (with default value) | ||
| + | ! scope="col" | Description | ||
| + | |- | ||
| + | | condLdColors="gray60,#E41A1C,#377EB8,#4DAF4A,#984EA3,#FF7F00,#A65628,#F781BF" | ||
| + | | First color is missing LD color, the rest are used as needed for each additional lead SNP | ||
| + | |- | ||
| + | | drawMarkerNames = T | ||
| + | | Display marker names (or not) above lead SNPs | ||
| + | |- | ||
| + | | condLdLow=NULL | ||
| + | | Used to set all SNPs with LD in the lowest bin to the same color, for example condLdLow="gray70" | ||
| + | |- | ||
| + | | condRefsnpPch=23 | ||
| + | | Symbol for each lead SNP, defaults to diamond | ||
| + | |- | ||
| + | | condPch='4,16,17,15,25,8,7,13,12,9,10' | ||
| + | | Plotting symbols for groups of SNPs in LD with additional refsnps, make sure they don't overlap with condRefsnpPch above | ||
| + | |- | ||
| + | | ldCuts = "0,.2,.4,.6,.8,1" | ||
| + | | Bins for LD | ||
| + | |} | ||
| + | |||
| + | === GWAS catalog variants === | ||
| + | |||
| + | You can add known GWAS variants to your plots. For example: | ||
| + | |||
| + | <syntaxhighlight lang="bash"> | ||
| + | locuszoom ... --gwas-cat whole-cat_significant-only --build hg19 | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | [[File:New lz gwas cat.png|900px]] | ||
| + | |||
| + | |||
| + | Currently the only catalog is the NHGRI GWAS catalog from [http://www.genome.gov/gwastudies/ genome.gov]. | ||
| + | |||
| + | <pre> | ||
| + | Available GWAS catalogs for build hg19: | ||
| + | |||
| + | +----------------------------+----------------------------------------------------------------+ | ||
| + | | Option | Description | | ||
| + | +----------------------------+----------------------------------------------------------------+ | ||
| + | | whole-cat_significant-only | The entire GWAS catalog, filtered to SNPs with p-value < 5E-08 | | ||
| + | +----------------------------+----------------------------------------------------------------+ | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | If the R package '''gridExtra''' is installed, a summary of each GWAS catalog variant in your region is listed later in the PDF: | ||
| + | |||
| + | [[File:New lz gwas summary.png|500px]] | ||
| + | |||
| + | === Fine-mapping credible sets === | ||
| + | |||
| + | LocusZoom can add an additional track to the plot showing results from a fine-mapping analysis. These are typically SNPs within the 95% credible set (see [http://www.nature.com/ng/journal/v44/n12/full/ng.2435.html this paper] for an example of a method generating such a set of SNPs.) | ||
| + | |||
| + | To add this fine-mapping track, you supply (as a plotting option) the fine-mapping set of credible SNPs as a file: | ||
| + | |||
| + | <syntaxhighlight lang="bash"> | ||
| + | locuszoom ... fineMap="my_finemapping_results.txt" | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | |||
| + | The fine-mapping results file should be a tab-delimited file with each fine-mapping SNP (for example, all those fine-mapping SNPs in the 95% credible set), a descriptive label (EUR/AMR/AFR/etc.), and a color: | ||
| + | |||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | ! scope="col" | snp | ||
| + | ! scope="col" | chr | ||
| + | ! scope="col" | pos | ||
| + | ! scope="col" | pp | ||
| + | ! scope="col" | group | ||
| + | ! scope="col" | color | ||
| + | |- | ||
| + | | rs1 || 18 || 55931115 || 0.88 || AMR || red | ||
| + | |- | ||
| + | | rs1 || 18 || 55920115 || 0.88 || AMR || red | ||
| + | |- | ||
| + | | rs1 || 18 || 55940115 || 0.88 || AMR || red | ||
| + | |- | ||
| + | | rs1 || 18 || 55930115 || 0.88 || EUR || blue | ||
| + | |- | ||
| + | | rs2 || 18 || 55940115 || 0.02 || EUR || blue | ||
| + | |- | ||
| + | | rs3 || 18 || 56000000 || 0.03 || AFR || green | ||
| + | |- | ||
| + | | rs4 || 18 || 56022000 || 0.03 || AFR || green | ||
| + | |- | ||
| + | | rs3 || 18 || 56100000 || 0.03 || ASN || purple | ||
| + | |- | ||
| + | | rs3 || 18 || 56150000 || 0.03 || ASN || purple | ||
| + | |- | ||
| + | | rs4 || 18 || 56160000 || 0.03 || ASN || purple | ||
| + | |- | ||
| + | | rs4 || 18 || 56180000 || 0.03 || ASN || purple | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | LocusZoom will extract from the file only those SNPs falling within the region to be plotted, so you can provide all of your fine-mapping results in a single file. | ||
| + | |||
| + | |||
| + | The generated plot will have a track showing the fine-mapping SNPs: | ||
| + | |||
| + | [[File:New lz finemap.png|900px]] | ||
| + | |||
| + | |||
| + | If the R package '''gridExtra''' is installed, the PDF will also have a summary of each fine-mapping SNP: | ||
| + | |||
| + | [[File:New lz finemap summary.png|400px]] | ||
| + | |||
| + | === Labeling multiple SNPs === | ||
| + | |||
| + | You can specify a file controlling the labels for either the reference SNP, or any other arbitrary SNP within the region. For example: | ||
| + | |||
| + | [[File:New lz denote markers.png|700px]] | ||
| + | |||
| + | Use the --denote-markers-file <file> argument to do this: | ||
| + | |||
| + | <syntaxhighlight lang="bash"> | ||
| + | locuszoom ... --denote-markers-file <your file> | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | The file looks like: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | ! scope="col" align="left" | snp | ||
| + | ! scope="col" align="left" | string | ||
| + | ! scope="col" align="left" | color | ||
| + | |- | ||
| + | | rs231362 || GWAS || blue | ||
| + | |- | ||
| + | | rs163184 || Conditional || purple | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | It must be tab-delimited and the columns must have a header and be named as such. | ||
| + | |||
| + | === Plotting BED tracks === | ||
| + | |||
| + | You can supply locuszoom with a BED file, and the tracks within it will be added to the plot. For example: | ||
| + | |||
| + | [[File:Bed_tracks.png]] | ||
| + | |||
| + | Use the --bed-tracks option, for example: | ||
| + | |||
| + | <pre> | ||
| + | locuszoom ... --bed-tracks <your bed file> | ||
| + | </pre> | ||
| + | |||
| + | The BED file should have at least 4 columns: the first 3 for chr/start/end, and the 4th column for the label of the track. It must be '''tab-delimited''', not white-space delimited. | ||
| + | |||
| + | Color can also be specified, but the BED file then needs to follow the full [http://genome.ucsc.edu/FAQ/FAQformat.html#format1 BED format]. | ||
| + | |||
| + | === Specify gene table (refFlat, GENCODE, etc.) === | ||
| + | |||
| + | You can now specify a different gene information table to use. LocusZoom provides both refFlat and GENCODE. refFlat is the default. For example: | ||
| + | |||
| + | <pre> | ||
| + | locuszoom --gene-table gencode | ||
| + | </pre> | ||
== Output == | == Output == | ||
| Line 347: | Line 727: | ||
| --markercol | | --markercol | ||
| Name of the SNP column in the --metal file. | | Name of the SNP column in the --metal file. | ||
| + | |- | ||
| + | | --epacts | ||
| + | | Provide a results file generated by [[EPACTS]] instead of a --metal file. | ||
|- | |- | ||
| --refsnp | | --refsnp | ||
| Line 357: | Line 740: | ||
| Specify the region near a reference SNP or gene as a "flank", instead of having to specify chr/start/stop explicitly. This can be specified in bases, kilobases, or megabases. Examples: 500kb, 1MB, 100141 | | Specify the region near a reference SNP or gene as a "flank", instead of having to specify chr/start/stop explicitly. This can be specified in bases, kilobases, or megabases. Examples: 500kb, 1MB, 100141 | ||
|- | |- | ||
| − | | --chr, --start, -- | + | | --chr, --start, --end |
| Specify chromosome/start/stop as the exact interval to plot. If no --refsnp is specified, the SNP with the most significant p-value in the region will be used as the reference SNP. | | Specify chromosome/start/stop as the exact interval to plot. If no --refsnp is specified, the SNP with the most significant p-value in the region will be used as the reference SNP. | ||
|- | |- | ||
| Line 363: | Line 746: | ||
|- | |- | ||
| --build | | --build | ||
| − | | Human genome build. This defaults to "hg18" | + | | Human genome build. This defaults to "hg18". You can supply your own build-specific data by modifying the conf file, and creating your own SQLite database (see *LINK HERE*). |
|- | |- | ||
| --ld | | --ld | ||
| Provide a file specifying LD between your reference SNP and all SNPs within the region you wish to plot. You only need to supply this file if you have created LD specifically for your purposes (perhaps a different population or genome build.) Otherwise, LD is computed automatically for you. | | Provide a file specifying LD between your reference SNP and all SNPs within the region you wish to plot. You only need to supply this file if you have created LD specifically for your purposes (perhaps a different population or genome build.) Otherwise, LD is computed automatically for you. | ||
| + | |- | ||
| + | | --ld-vcf | ||
| + | | Use a VCF file to calculate LD between SNPs. This can be a VCF file with an entire genome of SNPs and does not have to be subsetted to your region. The VCF file must also have a tabix index file. For calculating D', the VCF must be phased. | ||
|- | |- | ||
| --source | | --source | ||
| Line 375: | Line 761: | ||
|- | |- | ||
| --snpset | | --snpset | ||
| − | | Rug of SNPs to create at the top of the plot. Defaults to the Illumina 1M chip currently. | + | | Rug of SNPs to create at the top of the plot. Defaults to the Illumina 1M chip currently. To disable, use --snpset NULL. |
|- | |- | ||
| --plotonly | | --plotonly | ||
| Line 381: | Line 767: | ||
|- | |- | ||
| --no-transform | | --no-transform | ||
| − | | LocusZoom supports arbitrary precision p-values. However, if your p-values have already been transformed to the | + | | LocusZoom supports arbitrary precision p-values. However, if your p-values have already been transformed to the -log10 scale, you can use this option to stop LocusZoom from automatically transforming them. |
|- | |- | ||
| --prefix | | --prefix | ||
| Line 398: | Line 784: | ||
In addition to the options above, there are options that control the plotting engine inside Locuszoom. These are used with a different syntax: arg=value (no spaces allowed). | In addition to the options above, there are options that control the plotting engine inside Locuszoom. These are used with a different syntax: arg=value (no spaces allowed). | ||
| + | New/fixed options in 1.3: | ||
| + | |||
| + | {| width="85%" cellspacing="0" cellpadding="5" border="1" | ||
| + | |- | ||
| + | ! scope="col" | Option (with default value) | ||
| + | ! scope="col" | Description | ||
| + | |- | ||
| + | | colorCol=NULL | ||
| + | | Specify the name of a column in association results file denoting the color each marker should be. This disables coloring by LD. For the column values, color names should be used, for example "red" "olivedrab" etc. | ||
| + | |- | ||
| + | | signifLine=NULL | ||
| + | | Specify (in -log10 p-value scale) where to place a horizontal significance line. Can have multiple lines, e.g. signifLine="7.3,9" | ||
| + | |- | ||
| + | | signifLineColor=NULL | ||
| + | | Specify color of each significance line, e.g. signifLineColor="red,blue" | ||
| + | |- | ||
| + | | signifLineWidth=NULL | ||
| + | | Specify the line width for each significance line, e.g. signifLineWidth="2,3" | ||
| + | |- | ||
| + | | showIso=F | ||
| + | | Show genes as isoforms, rather than collapsed into one canonical transcript. To enable use showIso=T | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Other options: | ||
{| width="85%" cellspacing="0" cellpadding="5" border="1" | {| width="85%" cellspacing="0" cellpadding="5" border="1" | ||
|- | |- | ||
| Line 592: | Line 1,004: | ||
<code></code> | <code></code> | ||
<pre>--metal your_data --refsnp rs7983146 --flank 500kb | <pre>--metal your_data --refsnp rs7983146 --flank 500kb | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</pre> | </pre> | ||
=== Use 1000 genomes, CEU for LD instead of the default (HapMap r22 CEU) === | === Use 1000 genomes, CEU for LD instead of the default (HapMap r22 CEU) === | ||
| Line 607: | Line 1,014: | ||
<code></code> | <code></code> | ||
| − | <pre>--metal your_data --refsnp rs11899863 --pop YRI | + | <pre>--metal your_data --refsnp rs11899863 --pop YRI --build hg18 --source hapmap |
</pre> | </pre> | ||
| Line 613: | Line 1,020: | ||
<code></code> | <code></code> | ||
| − | <pre>--metal your_data --refsnp rs1552224 --chr 11 --start 71810746 -- | + | <pre>--metal your_data --refsnp rs1552224 --chr 11 --start 71810746 --end 72710746 |
</pre> | </pre> | ||
| − | === An example using plotting | + | === An example using plotting options === |
| + | |||
| + | Note in this example the plotting options are placed at the '''end''' of the command-line, and are of the format '''arg'''='''value'''. The value should be double-quoted if spaces are included in the value (see title= below.) | ||
<code></code> | <code></code> | ||
| Line 624: | Line 1,033: | ||
== Advanced configuration == | == Advanced configuration == | ||
| − | === Creating a SQLite database === | + | === Creating a custom SQLite database === |
| − | As a starting point, we provide | + | As a starting point, we provide SQLite databases based on UCSC human genome '''build hg18 and hg19''', which includes the following tables: |
*snp_pos: SNP positions | *snp_pos: SNP positions | ||
| Line 850: | Line 1,259: | ||
If you wish for your database to become the default, change the <code>LATEST_BUILD</code> variable in the m2zfast.conf file to whatever you have chosen above (in our example, our new database became mapped to 'hg19'.) | If you wish for your database to become the default, change the <code>LATEST_BUILD</code> variable in the m2zfast.conf file to whatever you have chosen above (in our example, our new database became mapped to 'hg19'.) | ||
| + | |||
| + | === Updating the existing locuszoom database(s) === | ||
| + | |||
| + | LocusZoom now comes with a database updating script <code>bin/lzupdate.py</code>. This script can download the necessary data from UCSC, NCBI, NGHRI, and GENCODE to create an up-to-date database file. The script performs the following actions: | ||
| + | |||
| + | # Download latest SNP table from UCSC for the given build | ||
| + | # Reformat SNP table for insertion into sqlite database | ||
| + | # Download latest refFlat from UCSC for the given build | ||
| + | # Reformat refFlat for insertion into sqlite database | ||
| + | # (optional) Download GENCODE annotation file from GENCODE FTP site | ||
| + | # Download RsMergeArch from NCBI | ||
| + | # Write formatted translation table for old rsIDs to latest (from RsMergeArch) | ||
| + | # Create a SNP set file (for indicating rug of markers at top of plot for different genotyping arrays) | ||
| + | # Download the latest NHGRI GWAS catalog | ||
| + | # Format catalog for use with locuszoom | ||
| + | # Call <code>bin/dbmeister.py</code> to insert everything above (except the GWAS catalog file, which remains a separate file) | ||
| + | |||
| + | An example of running the script: | ||
| + | |||
| + | <pre> | ||
| + | bin/lzupdate.py --build hg19 --gencode 19 --gwas-cat | ||
| + | </pre> | ||
| + | |||
| + | The script will NOT overwrite the existing locuszoom database, since you should likely back it up first (under data/database/*.db). After running the script you should have both a new locuszoom.db file, and a gwas catalog file. You can then either overwrite the locuszoom database after backing it up, or you could place them in a different location and modify the conf file accordingly. The script will provide instructions after running for how to do this. | ||
=== Changing m2zfast.conf settings === | === Changing m2zfast.conf settings === | ||
| Line 860: | Line 1,293: | ||
! scope="col" | Description | ! scope="col" | Description | ||
|- | |- | ||
| − | | | + | | DEFAULT_BUILD |
| − | | | + | | Default build to use for finding SNP positions, and calculating LD. This is used to index SQLITE_DB, as well as LD_DB. |
| + | |- | ||
| + | | DEFAULT_POP | ||
| + | | Default population to use for computing LD. | ||
| + | |- | ||
| + | | DEFAULT_SOURCE | ||
| + | | Default source for computing LD. | ||
|- | |- | ||
| NEWFUGUE_PATH | | NEWFUGUE_PATH | ||
| Path to the new_fugue binary. Defaults to "new_fugue", which simply means it is searched for on your path. If new_fugue is not on your path, you will need to specify the full path here. | | Path to the new_fugue binary. Defaults to "new_fugue", which simply means it is searched for on your path. If new_fugue is not on your path, you will need to specify the full path here. | ||
|- | |- | ||
| − | | | + | | PLINK_PATH |
| − | | | + | | Path to the PLINK binary. Defaults to "plink", which searches for PLINK on your path. If it is not on your path, specify the full path here. |
| + | |- | ||
| + | | RSCRIPT_PATH | ||
| + | | Path to the Rscript binary. Defaults to "Rscript", which searches for Rscript on your path. If it is not on your path, specify the full path here. | ||
|- | |- | ||
| SQLITE_DB | | SQLITE_DB | ||
| Line 875: | Line 1,317: | ||
| Contains a "tree" which maps a tuple of (genotype source, genotype population, genome build) to genotype files. | | Contains a "tree" which maps a tuple of (genotype source, genotype population, genome build) to genotype files. | ||
|- | |- | ||
| − | | | + | | GWAS_CATS |
| − | | | + | | Contains a "tree" which maps genome build and the name of a GWAS catalog to the actual file containing the GWAS hits. |
| − | |||
| − | |||
| − | |||
|} | |} | ||
Revision as of 13:00, 16 May 2018
This page contains information regarding a version of LocusZoom that may be downloaded for personal use. For more information on LocusZoom, see this page.
To be notified of future changes to LocusZoom, you can join our Google Group.
Quick Start (Requirements)
The following software is required:
- Python 2.7+ (do not download the 3.0 branch!)
- R 3.0+. Note that if using R 3.1, you must install LocusZoom 1.3 (previous versions will fail.)
The following software is optional but recommended:
- new_fugue, a program for computing LD, written by Goncalo Abecasis.
- PLINK
- tabix, downloaded with samtools
The following R packages are optional but recommended:
- gridExtra (used for creating summary tables of GWAS hits / fine-mapping SNPs as additional pages in the PDF)
For the latest stable LocusZoom package, see our download page. The current version is 1.3, released on June 20th, 2014.
Currently only Unix/Linux is supported, though Mac OS X should be supported in a future release. Support for Windows may come at a much later date.
Synopsis
First, change directory into examples/. Then, run the following command:
./run_example.py
This script runs the following command for you:
../bin/locuszoom --metal Kathiresan_2009_HDL.txt --refgene FADS1
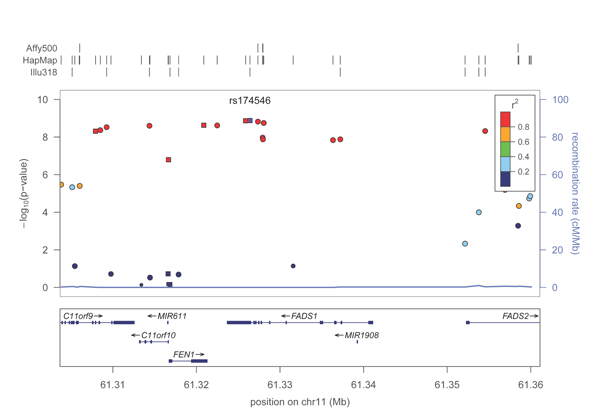
A PDF plot of the FADS1 locus will be created in the directory. It should look roughly like this:
Download
See our download page for links to the latest as well as previous releases.
Installation
Step 1: Install Python
You will need to install Python on your system if it is not already. Head over to www.python.org to download it. Note that you will want to make sure to download the latest from the 2.x branch, and *not* the 3.x one.
Step 2: Install R
R is also required for generating the plots. You can download R at www.r-project.org. Version 2.10 or greater is required.
Step 3: Install LD calculation software (optional)
- If you wish to calculate from hg18 sources (hapmap, earlier releases of 1000G): install new_fugue (see below.)
- If you wish to calculate from hg19 sources (latest 1000G): install PLINK (see below.)
- If you plan to supply your own LD files per region, or calculate LD directly from VCF files: install nothing! See options for --ld and --ld-vcf.
new_fugue
New_fugue is a program that calculates linkage disequilibrium measures from genotype files. While installing new_fugue is optional, we highly recommend it as it makes the process of generating plots much easier. If you opt to skip installing new_fugue, you will need to provide your own computed LD files for each region that you want to plot.
New_fugue can be downloaded from here.
Once downloaded, extract the tar file using:
tar zxf /path/to/new_fugue.tar.gz
Change into the generic-new_fugue directory that is created, and run:
make install
You may need administrator rights to install this program.
PLINK
PLINK is now used to calculate LD for all future LD sources / populations that we may add. The program new_fugue (above) is used to calculate LD from older sources (such as hapmap) and older builds (such as hg18) where LD files are sufficiently small.
You can download PLINK and find instructions for installing it here.
Step 5: Install tabix
Tabix is used to quickly extract regions from bgzipped and tabix-indexed files. It is used in LocusZoom to extract regions from VCF files when calculating LD, and for extracting from EPACTS result files.
It can be downloaded from the sourceforge site here or directly to the download site here.
Step 6: Install LocusZoom
LocusZoom is provided as a tar archive which contains the following:
- The LocusZoom python application
- The R script used for generating plots
- Human genome build hg18 and hg19 data, including:
- genotype files (used for computing LD) from HapMap and 1000G
- a SQLite database file containing tables describing SNP positions, SNP annotations, gene and exon locations, and recombination rates
Simply unpack the tar to your directory of choice by doing the following:
cd <directory where you want to place locuszoom> tar zxf /path/to/locuszoom.tgz
The tar archive will extract into the following directory structure:
- locuszoom/
- bin/
- locuszoom (this is the locuszoom "executable")
- locuszoom.R (the R script which is used by locuszoom for creating the plots)
- dbmeister.py (script for creating custom user databases)
- lzupdate.py (script for creating an updated copy of the provided locuszoom database)
- conf/ (configuration file located here)
- data/
- database/ (SQLite file located here)
- hapmap/ (hapmap genotype files)
- 1000G/ (1000G genotype files)
- src/ (source code for locuszoom)
- bin/
It is important that this directory structure remain intact. To make launching locusoom easier, you could create a link to it from /usr/local/bin, for example:
ln -s bin/locuszoom /usr/local/bin/locuszoom
Sources of information
LocusZoom uses various sources of information for annotation, positions, and calculating LD.
For computing LD:
- HapMap genotypes for populations CEU, YRI, and JPT+CHB.
- 1000 Genomes genotypes
For SNP, gene, and exon positions:
- dbSNP position via the UCSC Genome Browser
- Gene and exon positions via the UCSC Genome Browser. We filtered SNPs that map to more than one location or where no allele matches the reference sequence.
For annotation:
- We use various sources including RefSeq Genes (refFlat), TFBS Conserved (tfbsConsSites), and Conservation (phaseConsElements44wayPlacental), all available from the UCSC Genome Browser.
- Recombination rates from HapMap.
For GWAS hits:
- We use the NHGRI GWAS catalog, available at genome.gov
Input
Association results file
LocusZoom requires an association results file similar in formatting to what METAL or EPACTS provides.
METAL formatted file
The file must have 2 columns: markers (SNPs), and p-values. The file should look something like this:
| MarkerName | P-value |
|---|---|
| rs1 | 0.423 |
| rs2 | 1.23e-04 |
| chr4:401141 | 9.4e-390 |
Markers can be either rsIDs or chr:pos format (see above).
This file should be passed to locuszoom using the --metal option.
The file should be tab-delimited, though this can be changed using the --delim option.
If your marker and p-value column names are not "MarkerName" and "P-value", you may set them with --markercol and --pvalcol options.
P-values of any magnitude are supported in scientific notation (we use an arbitrary precision library built-in to python, and transform p-values to the log scale.) If you've already transformed your p-values to the log scale, simply use --no-transform and LocusZoom will not transform them.
EPACTS formatted file
The file can come directly from EPACTS, or simply be formatted similarly to the following:
| #CHROM | BEGIN | END | MARKER_ID | NS | AC | CALLRATE | MAF | PVALUE | SCORE | N.CASE | N.CTRL | AF.CASE | AF.CTRL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 15903 | 15903 | 1:15903_G/GC | 2657 | 3892.2 | 1 | 0.26757 | 0.36771 | 0.90077 | 1326 | 1331 | 1.4688 | 1.4609 |
| 1 | 19190 | 19191 | 1:19190_GC/G | 2657 | 823.65 | 1 | 0.155 | 0.67173 | 0.42378 | 1326 | 1331 | 0.3115 | 0.30849 |
| 1 | 20316 | 20317 | 1:20316_GA/G | 2657 | 1005.3 | 1 | 0.18917 | 0.50804 | 0.66189 | 1326 | 1331 | 0.38062 | 0.37607 |
| 1 | 30967 | 30970 | 1:30967_CCCA/C | 2657 | 435.35 | 1 | 0.081925 | 0.08848 | -1.7035 | 1326 | 1331 | 0.16007 | 0.16762 |
| 1 | 51972 | 51975 | 1:51972_GGAC/G | 2657 | 207.8 | 1 | 0.039104 | 0.51638 | -0.64893 | 1326 | 1331 | 0.077187 | 0.079226 |
| 1 | 53138 | 53140 | 1:53138_TAA/T | 2657 | 216.2 | 1 | 0.040685 | 0.55679 | 0.58762 | 1326 | 1331 | 0.083145 | 0.079602 |
| 1 | 54421 | 54421 | 1:54421_A/G | 2657 | 179.45 | 1 | 0.033769 | 0.73592 | 0.33726 | 1326 | 1331 | 0.068213 | 0.066867 |
| 1 | 66221 | 66221 | 1:66221_A/AT | 2657 | 664.45 | 1 | 0.12504 | 0.48676 | 0.69547 | 1326 | 1331 | 0.25366 | 0.24651 |
| 1 | 66222 | 66223 | 1:66222_TA/T | 2657 | 470.3 | 1 | 0.088502 | 0.64258 | 0.4641 | 1326 | 1331 | 0.17941 | 0.17461 |
The chrom, start, end, marker ID, and p-value columns must all be present. The file must be tab-delimited.
To load this file, use --epacts.
Note: LocusZoom (as of 1.3) will now use the tabix index for the EPACTS file if it exists and if tabix is intalled on your system. This results in much faster loading of EPACTS files and should absolutely be used if possible.
Warning: The "test" version of EPACTS changed the format of the output. To make LZ work, you'll also need to add --epacts-beg-col BEG to your command line.
Reading from STDIN
If you have a quick way of pulling out regions from your association results to plot (such as with tabix), you can pass the data directly to locuszoom on STDIN by specifying the file as "-". For example:
tabix -h my_file.gz 1:1-10000 | locuszoom --metal - --refgene TCF7L2
Region
You can specify the region to plot in any one of the following ways:
- A reference SNP and flanking region
--refsnp <your snp> --flank 500kb
- A reference SNP and chromosome/start/stop specification
--refsnp <your snp> --chr # --start <base position> --end <base position>
- A reference SNP and gene:
--refsnp <your snp> --refgene <your gene>
This will use the reference gene as the plotting boundaries. You can extend the boundaries by also including --flank.
- A gene and flanking region
--refgene <your gene> --flank 250kb
The flank is computed as +/- from the transcription start/end of the gene. From this region, LocusZoom will find the SNP with the most significant p-value, and use this as the reference SNP.
- A gene and chromosome/start/stop specification
--refgene <your gene> --chr # --start <base position> --end <base position>
This method is similar to the above, except that an exact region is specified. LD with the SNP with the most significant p-value in this region will be used to color data points.
- A chromosome/start/stop specification
--chr # --start <base position> --end <base position>
The SNP with the most significant p-value in this region will be used for estimating LD.
Specifying LD source/population/build
We supply genotype files for computing LD between the reference SNP and all other SNPs within the region you are plotting. The tables below show the supported combinations of LD source, population, and build. Note that you can always provide your own LD files, see User-supplied LD for more information.
1000G
| Release | Build | Population | LocusZoom Arguments |
|---|---|---|---|
| March 2012 | hg19 | ASN | --pop ASN --build hg19 --source 1000G_March2012 |
| March 2012 | hg19 | AFR | --pop AFR--build hg19 --source 1000G_March2012 |
| March 2012 | hg19 | EUR | --pop EUR --build hg19 --source 1000G_March2012 |
| March 2012 | hg19 | AMR | --pop AMR --build hg19 --source 1000G_March2012 |
| Nov 2010 | hg19 | ASN | --pop ASN --build hg19 --source 1000G_Nov2010 |
| Nov 2010 | hg19 | AFR | --pop AFR --build hg19 --source 1000G_Nov2010 |
| Nov 2010 | hg19 | EUR | --pop EUR --build hg19 --source 1000G_Nov2010 |
| June 2010 | hg18 | CEU | --pop CEU --build hg18 --source 1000G_June2010 |
| June 2010 | hg18 | YRI | --pop YRI --build hg18 --source 1000G_June2010 |
| June 2010 | hg18 | JPT+CHB | --pop JPT+CHB --build hg18 --source 1000G_June2010 |
| August 2009 | hg18 | CEU | --pop CEU --build hg18 --source 1000G_Aug2009 |
| August 2009 | hg18 | YRI | --pop YRI --build hg18 --source 1000G_Aug2009 |
| August 2009 | hg18 | JPT+CHB | --pop JPT+CHB --build hg18 --source 1000G_Aug2009 |
HapMap Phase II
| Build | Population | LocusZoom Arguments |
|---|---|---|
| hg18 | CEU | --pop CEU --build hg18 --source hapmap |
| hg18 | YRI | --pop YRI--build hg18 --source hapmap |
| hg18 | JPT+CHB | --pop JPT+CHB --build hg18 --source hapmap |
Batch mode
LocusZoom provides a batch mode for generating plots for a large number of regions:
--hitspec, which reads a batch mode specification file.
For this option, you must provide a text file of the following format:
| Column | Description |
|---|---|
| snp | Can be either a SNP, or gene. |
| chr | Chromosome |
| start | Start position to display on plot. |
| end | End position to display on plot. |
| flank | Flank for region. Can be given instead of chr/start/stop. |
| run | Should this row be read? Should be "yes" or "no". |
| m2zargs | List of arguments for customizing plots. You can find a list of them here: Commonly Used LocusZoom Options |
The file should be delimited by whitespace (tab, space, multiple spaces), and the header must exist, with column names exactly as specified in the table above. As an example, consider the following file:
| snp | chr | start | stop | flank | run | m2zargs |
|---|---|---|---|---|---|---|
| rs7983146 | NA | NA | NA | 500kb | yes | title="My favorite SNP" |
| TCF7L2 | NA | NA | NA | 1.25MB | yes | title="TCF7L2 Region" showRecomb=F |
| rs7957197 | 12 | 119503590 | 120322280 | NA | yes | showAnnot=F |
The first row would plot rs7983146 as the reference SNP, and a region of 500kb on either side of it. The plot title would read "My favorite SNP."
The second row would plot 1.25 MB on either side of TCF7L2's transcription start and stop. The SNP with the most significant p-value in your --metal file will be used as the reference SNP. The plot title would read "TCF7L2 Region", and the recombination overlay would be disabled using showRecomb=F.
The third row would plot rs7957197 as the reference SNP, but here we've specifically designated the region to plot, which is chr12:119503590-120322280. We've also disabled showing SNP annotations with showAnnot=F.
User-supplied LD
If new_fugue is installed, LocusZoom will automatically compute LD between the reference SNP and all other SNPs within each region to be plotted. However, you may wish to provide your own file with LD information. This can be done with the --ld option, which requires a file of the following format:
| Column | Description |
|---|---|
| snp1 | Any SNP in your plotting region. |
| snp2 | Should always be the reference SNP in the region. |
| dprime | D' between snp2 (reference SNP) and snp1. |
| rsquare | r2 between snp2 (reference SNP) and snp1. |
The dprime column can be all missing if it is not known. Rsquare must be present, and must be valid data.
The file should be whitespace delimited, and the header (column names shown above) must exist.
Supply VCF files for calculating LD
You can give LocusZoom a VCF file directly to use for calculating LD:
locuszoom --ld-vcf my_genotypes.vcf.gz ...
This option takes the place of having to supply per-region pre-calculated LD (--ld) or having to specify --pop and --source for calculating LD from genotype files supplied by LZ.
Warning: The VCF file must also have a tabix index located in the same directory. For the above example, the tabix index "my_genotypes.vcf.gz.tbi" must exist.
You can also calculate D' from phased VCF files:
locuszoom --ld-vcf my_genotypes.vcf.gz --ld-measure dprime ...
The default measure is "rsquared".
In version 1.3, if you have VCF files separated out by chromosome, you can create a JSON file mapping chromosome name --> VCF file, and provide the JSON file to --ld-vcf. For example, the JSON file could look like:
{
"X": "/path/to/X.vcf.gz",
"Y": "/path/to/Y.vcf.gz",
"MT": "/path/to/MT.vcf.gz",
}
And then pass it directly using locuszoom --ld-vcf my_vcfs.json.
Optional Input
Plotting LD with additional reference SNPs
LocusZoom can now show LD with multiple SNPs in a region (for example, you might want to show LD with a number of SNPs from a conditional analysis.)
You give LocusZoom the usual reference SNP (used for centering the plot and calculating the region) but an additional set of lead/reference SNPs as well.
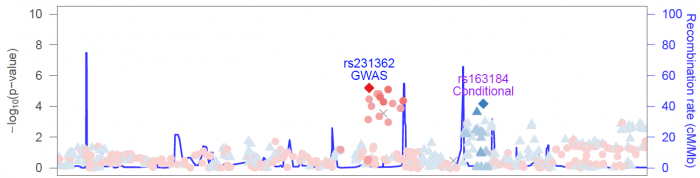
For all other SNPs not in the "lead SNP set" of { reference SNP, additional reference SNPs }, LZ will find which of the lead SNPs it is in highest LD with, and color it to match that lead SNP. The extent of LD with the lead SNP is shown by a gradient of color.
As an example:
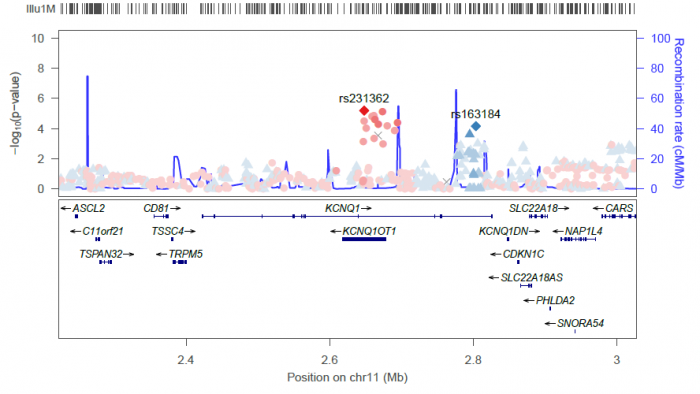
locuszoom --metal <DIAGRAM T2D results> --refsnp "rs231362" --add-refsnps "rs163184"
Will generate the following plot:
The following options are available for changing the style of these types of plots:
| Option (with default value) | Description |
|---|---|
| condLdColors="gray60,#E41A1C,#377EB8,#4DAF4A,#984EA3,#FF7F00,#A65628,#F781BF" | First color is missing LD color, the rest are used as needed for each additional lead SNP |
| drawMarkerNames = T | Display marker names (or not) above lead SNPs |
| condLdLow=NULL | Used to set all SNPs with LD in the lowest bin to the same color, for example condLdLow="gray70" |
| condRefsnpPch=23 | Symbol for each lead SNP, defaults to diamond |
| condPch='4,16,17,15,25,8,7,13,12,9,10' | Plotting symbols for groups of SNPs in LD with additional refsnps, make sure they don't overlap with condRefsnpPch above |
| ldCuts = "0,.2,.4,.6,.8,1" | Bins for LD |
GWAS catalog variants
You can add known GWAS variants to your plots. For example:
locuszoom ... --gwas-cat whole-cat_significant-only --build hg19
Currently the only catalog is the NHGRI GWAS catalog from genome.gov.
Available GWAS catalogs for build hg19: +----------------------------+----------------------------------------------------------------+ | Option | Description | +----------------------------+----------------------------------------------------------------+ | whole-cat_significant-only | The entire GWAS catalog, filtered to SNPs with p-value < 5E-08 | +----------------------------+----------------------------------------------------------------+
If the R package gridExtra is installed, a summary of each GWAS catalog variant in your region is listed later in the PDF:
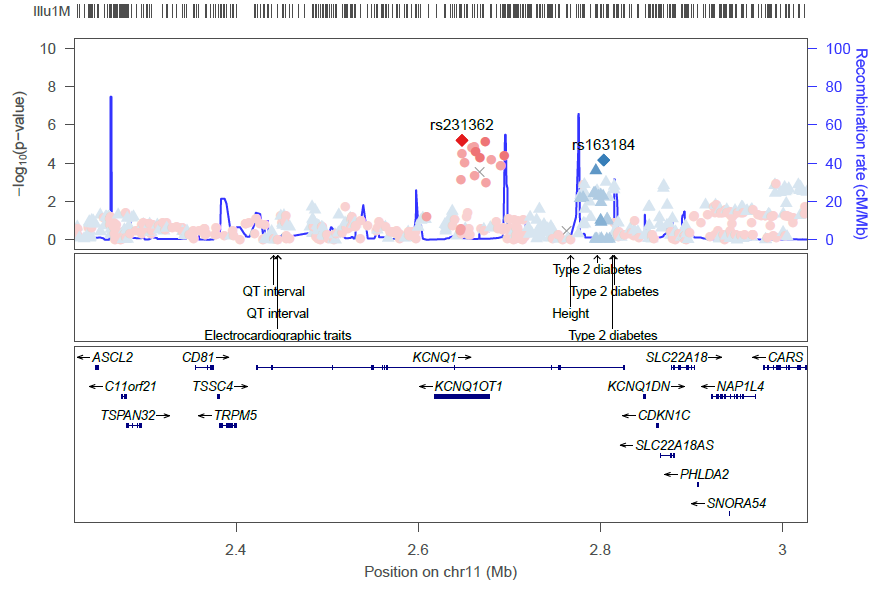
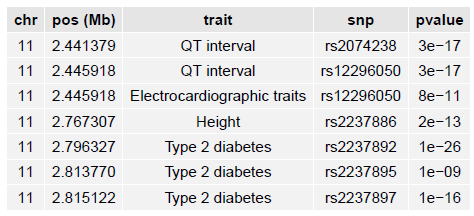
Fine-mapping credible sets
LocusZoom can add an additional track to the plot showing results from a fine-mapping analysis. These are typically SNPs within the 95% credible set (see this paper for an example of a method generating such a set of SNPs.)
To add this fine-mapping track, you supply (as a plotting option) the fine-mapping set of credible SNPs as a file:
locuszoom ... fineMap="my_finemapping_results.txt"
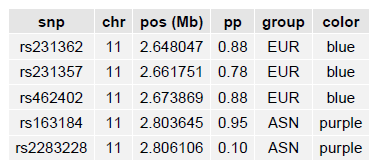
The fine-mapping results file should be a tab-delimited file with each fine-mapping SNP (for example, all those fine-mapping SNPs in the 95% credible set), a descriptive label (EUR/AMR/AFR/etc.), and a color:
| snp | chr | pos | pp | group | color |
|---|---|---|---|---|---|
| rs1 | 18 | 55931115 | 0.88 | AMR | red |
| rs1 | 18 | 55920115 | 0.88 | AMR | red |
| rs1 | 18 | 55940115 | 0.88 | AMR | red |
| rs1 | 18 | 55930115 | 0.88 | EUR | blue |
| rs2 | 18 | 55940115 | 0.02 | EUR | blue |
| rs3 | 18 | 56000000 | 0.03 | AFR | green |
| rs4 | 18 | 56022000 | 0.03 | AFR | green |
| rs3 | 18 | 56100000 | 0.03 | ASN | purple |
| rs3 | 18 | 56150000 | 0.03 | ASN | purple |
| rs4 | 18 | 56160000 | 0.03 | ASN | purple |
| rs4 | 18 | 56180000 | 0.03 | ASN | purple |
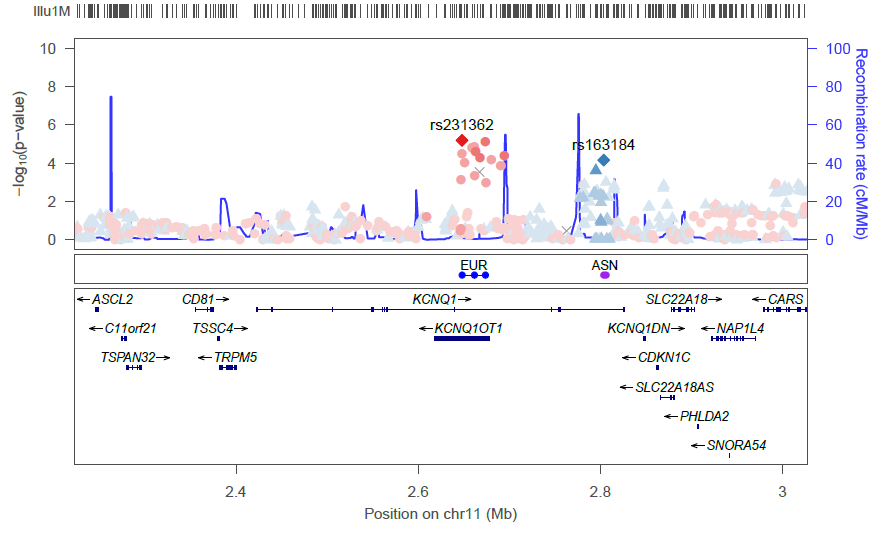
LocusZoom will extract from the file only those SNPs falling within the region to be plotted, so you can provide all of your fine-mapping results in a single file.
The generated plot will have a track showing the fine-mapping SNPs:
If the R package gridExtra is installed, the PDF will also have a summary of each fine-mapping SNP:
Labeling multiple SNPs
You can specify a file controlling the labels for either the reference SNP, or any other arbitrary SNP within the region. For example:
Use the --denote-markers-file <file> argument to do this:
locuszoom ... --denote-markers-file <your file>
The file looks like:
| snp | string | color |
|---|---|---|
| rs231362 | GWAS | blue |
| rs163184 | Conditional | purple |
It must be tab-delimited and the columns must have a header and be named as such.
Plotting BED tracks
You can supply locuszoom with a BED file, and the tracks within it will be added to the plot. For example:
Use the --bed-tracks option, for example:
locuszoom ... --bed-tracks <your bed file>
The BED file should have at least 4 columns: the first 3 for chr/start/end, and the 4th column for the label of the track. It must be tab-delimited, not white-space delimited.
Color can also be specified, but the BED file then needs to follow the full BED format.
Specify gene table (refFlat, GENCODE, etc.)
You can now specify a different gene information table to use. LocusZoom provides both refFlat and GENCODE. refFlat is the default. For example:
locuszoom --gene-table gencode
Output
LocusZoom will produce a directory for each plot that contains the plot itself, along with a number of temporary files containing information on your particular region. The plot will be a PDF, named with the chr#:start-stop that was plotted.
If you only want the PDF itself, and don't want the other files, you can use the --plotonly option.
Each directory (or PDF, in the case of --plotonly) will have the date included to avoid collisions with previous plots - this behavior can be disabled using --no-date.
You can further customize the directory/PDF names that are created by using the --prefix <name> option. This will append a text string at the beginning of each directory/PDF that is created.
LocusZoom options
LocusZoom has a number of command line options, described in the table below.
| Option | Description |
|---|---|
| Important settings | |
| --metal | This is the data file to provide. Files generated by the meta-analysis program METAL are already formatted appropriately. If your data is not from METAL, it is very simple to format it (see Input.) |
| --delim | Delimiter for the data file. This defaults to tab, but can be anything. For ease of specification, you can use the following shortcuts: --delim tab, --delim space, --delim comma. |
| --pvalcol | Name of p-value column in the --metal file. |
| --markercol | Name of the SNP column in the --metal file. |
| --epacts | Provide a results file generated by EPACTS instead of a --metal file. |
| --refsnp | Reference SNP to be used in the plot. |
| --refgene | Specify a gene instead of a reference SNP. This will plot a region near a gene, and automatically find the SNP with the most significant p-value to use as the reference SNP. |
| --flank | Specify the region near a reference SNP or gene as a "flank", instead of having to specify chr/start/stop explicitly. This can be specified in bases, kilobases, or megabases. Examples: 500kb, 1MB, 100141 |
| --chr, --start, --end | Specify chromosome/start/stop as the exact interval to plot. If no --refsnp is specified, the SNP with the most significant p-value in the region will be used as the reference SNP. |
| Optional settings | |
| --build | Human genome build. This defaults to "hg18". You can supply your own build-specific data by modifying the conf file, and creating your own SQLite database (see *LINK HERE*). |
| --ld | Provide a file specifying LD between your reference SNP and all SNPs within the region you wish to plot. You only need to supply this file if you have created LD specifically for your purposes (perhaps a different population or genome build.) Otherwise, LD is computed automatically for you. |
| --ld-vcf | Use a VCF file to calculate LD between SNPs. This can be a VCF file with an entire genome of SNPs and does not have to be subsetted to your region. The VCF file must also have a tabix index file. For calculating D', the VCF must be phased. |
| --source | Source to use for genotypes when using LD. See Specifying LD source/population/build for more info. |
| --pop | Population to use when computing LD. See Specifying LD source/population/build for more info. |
| --snpset | Rug of SNPs to create at the top of the plot. Defaults to the Illumina 1M chip currently. To disable, use --snpset NULL. |
| --plotonly | Create only a PDF of the plot, and remove all temporary files/directories created during plotting. |
| --no-transform | LocusZoom supports arbitrary precision p-values. However, if your p-values have already been transformed to the -log10 scale, you can use this option to stop LocusZoom from automatically transforming them. |
| --prefix | Places a text string at the beginning of each plot or directory created. This is mainly used to denote different batches of plots - for example, you could use --prefix using_ceu to denote these plots are computed using CEU LD information. |
| --db | SQLite database file to use. This is set in the conf file by default, but can be changed on the command line if desired. |
Plotting options
In addition to the options above, there are options that control the plotting engine inside Locuszoom. These are used with a different syntax: arg=value (no spaces allowed).
New/fixed options in 1.3:
| Option (with default value) | Description |
|---|---|
| colorCol=NULL | Specify the name of a column in association results file denoting the color each marker should be. This disables coloring by LD. For the column values, color names should be used, for example "red" "olivedrab" etc. |
| signifLine=NULL | Specify (in -log10 p-value scale) where to place a horizontal significance line. Can have multiple lines, e.g. signifLine="7.3,9" |
| signifLineColor=NULL | Specify color of each significance line, e.g. signifLineColor="red,blue" |
| signifLineWidth=NULL | Specify the line width for each significance line, e.g. signifLineWidth="2,3" |
| showIso=F | Show genes as isoforms, rather than collapsed into one canonical transcript. To enable use showIso=T |
Other options:
| Option (with default value) | Description |
|---|---|
| theme=NULL | Select a theme. A theme is a collection of other settings. Options include theme=publication and theme=black. |
| ymax=10 | the display range for log10(p-value) will be at least ymax (extended as necessary to avoid clipping) |
| axisSize=1 | scaling factor for axes |
| axisTextSize=1 | sclaing factor for axis labels |
| axisTextColor=gray30 | color of axis labels |
| refsnpTextColor=black | color for reference SNP label (use 'transparent' to hide this label) |
| refsnpTextSize=1 | scaling factor for reference SNP text size |
| refsnpTextAlpha=1 | transparency level for reference SNP label (1=opaque,0=transparent) |
| title = "" | title for plot |
| titleColor=black | color for title |
| width=10 | width of pdf (inches) |
| height=7 | height of pdf (inches) |
| leftMarginLines=5 | margin (in lines) on left |
| rightMarginLines=5 | margin (in lines) on right |
| unit=1000000 | bp per unit displayed in plot |
| showAnnot=TRUE | show annotation for each snp? |
| showGenes=TRUE | show genes? |
| annotCol='annotation' | column to use for custom annotation, if it exists |
| annotPch='24,24,25,22,22,8,7,21' | plot symbols for annotation |
| annotOrder=NULL | ordering of custom annotation classes (comma-separated list annotation strings in order, alphabetical by default) |
| showRefsnpAnnot=TRUE | show annotation for reference snp too? |
| ld=NULL | file for LD information |
| ldCuts="0,.2,.4,.6,.8,1" | cut points for LD coloring |
| ldColors="gray50,navy, lightskyblue,green, orange,red,purple3" | colors for LD |
| ldCol='rsquare' | name for LD column |
| LDTitle=NULL | title for LD legend |
| smallDot=.4 | smallest p-value cex |
| largeDot=.8 | largest p-value cex |
| refDot=NULL | largest p-value cex |
| rfrows=4 | max number of rows used for displaying genes |
| showPartialGenes=TRUE | should genes that don't fit completely be displayed? |
| geneFontSize=.8 | size for gene names |
| geneColor="navy" | color for genes |
| snpsetFile=NULL | use this file for SNPset rug data |
| rugColor=gray30 | color for snpset rugs |
| rugAlpha=1 | alpha for snpset rugs |
| metalRug=NULL | if not null, use as label for rug of metal positions |
| showRecomb=TRUE | show recombination rate? |
| recombColor=blue | color for recombination rate on plot |
| recombAxisColor=NULL | color for recombination rate axis labeing (default matches recombColor) |
| recombAxisAlpha=NULL | color for recombination rate axis labeing |
| recombOver=FALSE | overlay recombination rate? (else underlay it) |
| recombFill=FALSE | fill recombination rate? (else line only) |
| recombFillAlpha=0.2 | recomb fill alpha |
| recombLineAlpha=0.8 | recomb line/text alpha |
| frameColor=gray30 | frame color for plots |
| frameAlpha=1 | frame alpha for plots |
| legendSize=.8 | scaling factor of legend |
| legendAlpha=1 | transparency of legend background |
| legend='auto' | legend? (auto, left, right, or none) |
| hiStart=0 | start of highlighted region |
| hiEnd=0 | end of highlighted region |
| hiColor=blue | color used for highlighting |
| hiAlpha=0.1 | transparency level for highlighting |
| prelude=NULL | R code to execute after data is read but before plot is made (allows data modification) |
| postlude=NULL, | R code to execute after plot is made |
Examples
A quick peek at a particular SNP
--metal your_data --refsnp rs1002227
A quick peek at a particular gene
--metal your_data --refgene CETP
Plot 500kb on either side of a SNP
--metal your_data --refsnp rs7983146 --flank 500kb
Use 1000 genomes, CEU for LD instead of the default (HapMap r22 CEU)
--metal your_data --refsnp rs11899863 --source 1000G
Use HapMap YRI for LD
--metal your_data --refsnp rs11899863 --pop YRI --build hg18 --source hapmap
Specify a specific region and reference SNP to plot
--metal your_data --refsnp rs1552224 --chr 11 --start 71810746 --end 72710746
An example using plotting options
Note in this example the plotting options are placed at the end of the command-line, and are of the format arg=value. The value should be double-quoted if spaces are included in the value (see title= below.)
--metal your_data --refsnp rs7903146 title="My region" geneFontSize=1.1 recombColor="gray"
Advanced configuration
Creating a custom SQLite database
As a starting point, we provide SQLite databases based on UCSC human genome build hg18 and hg19, which includes the following tables:
- snp_pos: SNP positions
- refFlat: gene information (exons, transcription start/stops, etc.)
- recomb_rate: recombination rates from hapmap phase 2
- snp_set: maps each SNP to a "set" - for example, all SNPs on the Illumina 1M chip
- refsnp_trans: a table that maps SNPs from previous builds to the current build
To create your own database, we provide a script bin/dbmeister.py that can insert these tables for you. We recommend creating your own database file, rather than inserting tables into the default LocusZoom database. This script is capable of using python's built-in sqlite support, but for faster insertion of tables (about 2x faster), we recommend installing sqlite3 from www.sqlite.org.
Inserting snp_pos
First, create a file that looks like the following:
| snp | chr | pos |
|---|---|---|
| rs38343 | 1 | 93919141 |
| rs918141 | 7 | 763263 |
| chr4:9181 | 4 | 9181 |
The file should be: tab-delimited, must have a header, and the columns should be exactly in that order.
Now, you can create your own database, and insert this file by using:
dbmeister.py --db my_database.db --snp_pos my_snp_pos_file
This command creates a database called "my_database.db" and inserts the SNP position table into it. If "my_database.db" had existed already, it would drop the snp_pos table in it, and insert yours in its place.
One special note about adding SNP position tables: a refsnp_trans table will automatically be created for you, where each SNP maps to itself. If you have a list of SNPs from previous builds that you would like to map to a SNP in the current build, you can then insert your own refsnp_trans table (see below for more information on this table.)
Inserting refsnp_trans
The refsnp_trans table looks like the following:
| rs_orig | rs_current |
|---|---|
| rs840 | rs715 |
| rs1086 | rs940 |
| rs1234 | rs1067 |
The first column contains SNP names from older genome builds, and the rs_current column contains SNP names from the current genome build (i.e., the build your database file is anchored to.)
Inserting this table into your database is simply then:
dbmeister.py --db my_database.db --trans my_snp_translations_file
You will want to execute this command AFTER inserting the snp_pos table, since that command drops the existing translation table.
Inserting refFlat
The refFlat table mirrors what is currently supplied by the refFlat table in the UCSC database. The file should look like:
| geneName | name | chrom | strand | txStart | txEnd | cdsStart | cdsEnd | exonCount | exonStarts | exonEnds |
|---|---|---|---|---|---|---|---|---|---|---|
| EFCAB1 | NM_024593 | chr8 | - | 49798505 | 49810423 | 49799853 | 49810263 | 6 | 49798505, | 49799913, |
| HECTD3 | NM_024602 | chr1 | - | 45240806 | 45249614 | 45241750 | 45249516 | 21 | 45240806,45241927, | 45241835,45241999, |
| PTPN20B | NM_001042361 | chr10 | - | 48357047 | 48447587 | 48358657 | 48447532 | 8 | 48357047,48359393,48391320, | 48358692,48359456,48391411, |
You can insert this table into the database using:
dbmeister.py --db my_database.db --refflat my_refflat_file
Inserting recomb_rate
The recomb_rate table mirrors what is available from HapMap. The format is:
| chr | pos | recomb | cm_pos |
|---|---|---|---|
| 1 | 72434 | 0.0015 | 0 |
| 1 | 78032 | 0.0015 | 8.397e-06 |
| 1 | 554461 | 0.0015 | 0.00072304 |
| 1 | 554484 | 0.0015 | 0.000723075 |
| 1 | 555296 | 0.0015 | 0.000724293 |
| 1 | 558185 | 0.0015 | 0.000728627 |
The table can be inserted using:
dbmeister.py --db my_database.db --recomb_rate my_recomb_rate_file
Inserting snp_set
The snp_set table simply carries a mapping of SNPs to a particular set they may belong to. The table looks like:
| snp | snp_set |
|---|---|
| rs1000 | Illu1M |
| rs1000 | Illu1M |
| rs1000 | Illu1M |
| rs1000000 | Illu1M |
| rs10000000 | Illu1M |
| rs10000009 | Illu1M |
The first column is a SNP, and the second is the name of the set it belongs to. If a SNP belongs to multiple sets, you can duplicate that SNP multiple times, one for each set.
Inserting the table into your database can be done using:
dbmeister.py --db my_database.db --snp_set my_snpset_file
Making LocusZoom aware of your new database
Now that you've created your own database file, you need to make LocusZoom aware that it exists. There are two ways to do this:
- Edit conf/m2zfast.conf, and change the
SQLITE_DBvariable - Supply the
--dbcommand line option when invokingbin/locuszoom
Editing the m2zfast.conf file entails changing the following block of code:
SQLITE_DB = {
'hg18' : "data/database/locuszoom_hg18.db"
};
Here, we've mapped build "hg18" to the default database file that comes with LocusZoom. You can mimic this format and insert your own, for example:
SQLITE_DB = {
'hg18' : "data/database/locuszoom_hg18.db",
'hg19' : "data/database/my_database.db"
};
The location of your database should be either an absolute path to your file, or a path relative to the locuszoom/ root (like those seen above.)
If you wish for your database to become the default, change the LATEST_BUILD variable in the m2zfast.conf file to whatever you have chosen above (in our example, our new database became mapped to 'hg19'.)
Updating the existing locuszoom database(s)
LocusZoom now comes with a database updating script bin/lzupdate.py. This script can download the necessary data from UCSC, NCBI, NGHRI, and GENCODE to create an up-to-date database file. The script performs the following actions:
- Download latest SNP table from UCSC for the given build
- Reformat SNP table for insertion into sqlite database
- Download latest refFlat from UCSC for the given build
- Reformat refFlat for insertion into sqlite database
- (optional) Download GENCODE annotation file from GENCODE FTP site
- Download RsMergeArch from NCBI
- Write formatted translation table for old rsIDs to latest (from RsMergeArch)
- Create a SNP set file (for indicating rug of markers at top of plot for different genotyping arrays)
- Download the latest NHGRI GWAS catalog
- Format catalog for use with locuszoom
- Call
bin/dbmeister.pyto insert everything above (except the GWAS catalog file, which remains a separate file)
An example of running the script:
bin/lzupdate.py --build hg19 --gencode 19 --gwas-cat
The script will NOT overwrite the existing locuszoom database, since you should likely back it up first (under data/database/*.db). After running the script you should have both a new locuszoom.db file, and a gwas catalog file. You can then either overwrite the locuszoom database after backing it up, or you could place them in a different location and modify the conf file accordingly. The script will provide instructions after running for how to do this.
Changing m2zfast.conf settings
The m2zfast.conf configuration file contains a number of settings that are typically static, but could require user configuration. The table below lists each variable, and its purpose.
| Variable | Description |
|---|---|
| DEFAULT_BUILD | Default build to use for finding SNP positions, and calculating LD. This is used to index SQLITE_DB, as well as LD_DB. |
| DEFAULT_POP | Default population to use for computing LD. |
| DEFAULT_SOURCE | Default source for computing LD. |
| NEWFUGUE_PATH | Path to the new_fugue binary. Defaults to "new_fugue", which simply means it is searched for on your path. If new_fugue is not on your path, you will need to specify the full path here. |
| PLINK_PATH | Path to the PLINK binary. Defaults to "plink", which searches for PLINK on your path. If it is not on your path, specify the full path here. |
| RSCRIPT_PATH | Path to the Rscript binary. Defaults to "Rscript", which searches for Rscript on your path. If it is not on your path, specify the full path here. |
| SQLITE_DB | See Making LocusZoom aware of your database. |
| LD_DB | Contains a "tree" which maps a tuple of (genotype source, genotype population, genome build) to genotype files. |
| GWAS_CATS | Contains a "tree" which maps genome build and the name of a GWAS catalog to the actual file containing the GWAS hits. |
LD caching
LocusZoom attempts to remember LD calculations that were made on previous runs of the program to avoid having to re-calculate the same regional LD for subsequent runs. The process works as follows:
- For a given reference SNP and chr/start/stop:
- If LD has not been previously computed, use new_fugue to compute LD with the reference SNP and all other SNPs in the region, and store this result to the LD cache
- Else, retrieve the previously stored LD results
The cache intelligently stores LD from separate sources (hapmap, 1000G), populations, builds, and even different versions of genotype files separately.
Upon running LocusZoom, a file called "ld_cache.db" will automatically be created in the current directory, and LD computations will be stored there. If you wish to change the location of the LD cache, use --cache <file>. If you wish to disable LD caching, you can use --cache None.
Python note: The ld_cache.db is actually a shelve, and you can explore its contents using:
import shelve
d = shelve.open("ld_cache.db")
License
Copyright 2010 Ryan Welch, Randall Pruim
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.