Difference between revisions of "RAREMETAL Documentation"
Shuang Feng (talk | contribs) |
(Update contact address) |
||
| (413 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Category:RAREMETAL]] | |
| + | == Useful Wiki Pages == | ||
| − | + | * Git hub page: https://github.com/statgen/Raremetal | |
| + | * The [[RAREMETAL_Change_Log | Change Log]] | ||
| + | * The [[RAREMETAL_DOWNLOAD_%26_BUILD | DOWNLOAD page]] | ||
| + | |||
| + | * The [[Tutorial:_RAREMETAL|RAREMETAL Quick Start Tutorial]] | ||
| + | * The [[RAREMETAL METHOD]] | ||
| + | |||
| + | * The [[RAREMETAL FAQ]] | ||
| + | |||
| + | * The [[RAREMETAL Command Reference]] | ||
| + | |||
| + | * The [[RAREMETALWORKER|RAREMETALWORKER documentation]] | ||
| + | |||
| + | The [http://genome.sph.umich.edu/wiki/Rvtests '''rvtests'''] tool for rare-variant association analysis can also generate output compatible with RAREMETAL. | ||
| + | |||
| + | == Brief Description == | ||
| + | |||
| + | '''RAREMETAL''' is a computationally efficient tool for meta-analysis of rare variants using sequencing or genotyping array data. It takes summary statistics and LD matrices generated by [[Rare-Metal-Worker|'''RAREMETALWORKER''']] or [http://genome.sph.umich.edu/wiki/Rvtests '''rvtests'''], handles related and unrelated individuals, and supports both single variant and burden meta-analysis. It generates high quality plots by default and has options that allow users to build reports at different levels. | ||
| + | |||
| + | '''RAREMETAL''' is developed by Shuang Feng, Dajiang Liu and Gonçalo Abecasis. A R-package written by Dajiang Liu using the same methodology is [[RareMetals|'''available''']]. | ||
== Key Features == | == Key Features == | ||
| − | ''' | + | '''RAREMETAL''' has the following features: |
| − | * | + | * Performs gene-based or region-based meta analysis using Burden tests with the following methods: CMC_counts, Madsen-Browning, SKAT, and Variable Threshold. |
| − | * | + | * Performs single variant metal-analysis by default. |
| − | * | + | * Allows customized groups of variants to be tested. |
| − | * | + | * Allows conditional analysis to be performed in both gene-level meta-analysis and single variants meta-analysis. |
| − | * | + | * Generate QQ plots and manhattan plots by default. |
| + | |||
| + | == Approach == | ||
| − | + | The key idea behind meta-analysis with RAREMETAL is that various gene-level test statistics can be reconstructed from single variant score statistics and that, when the linkage disequilibrium relationships between variants are known, the distribution of these gene-level statistics can be derived and used to evaluate signifi-cance. Single variant statistics are calculated using the Cochran-Mantel-Haenszel method. Our method has been published in [http://www.nature.com/ng/journal/v46/n2/abs/ng.2852.html '''Liu et. al'''] in Nature Genetics. Please go to [http://genome.sph.umich.edu/wiki/RAREMETAL_method '''method'''] for details. | |
| − | |||
| − | |||
| − | == | + | == Download and Installation == |
| − | |||
| − | + | We have tested compilation using our source code on several platforms including Linux, and Mac OS X. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | For source code and executables together with instructions of building from source, please go to [[RAREMETAL_DOWNLOAD_%26_BUILD |'''DOWNLOAD source and executables''']]. | |
| − | |||
| − | |||
| − | |||
| − | + | For questions about compilation, please go to [[RAREMETAL_FAQ | '''FAQ''']]. | |
| − | === | + | == Basic Usage Instructions == |
| − | |||
| − | + | '''RAREMETAL''' is a command line tool. It is typically run from a Linux or Unix prompt by invoking the command <code>raremetal</code>. In the following are descriptions of basic usage for meta analysis. A detailed [[Tutorial:_RareMETAL|'''TUTORIAL''']] with toy data are also available. | |
| − | |||
| − | |||
| − | |||
| − | ==== | + | ==== Prepare Input Files==== |
| − | + | '''RAREMETAL''' requires the following basic input files: summary statistics and covariance matrices of score statistics generated by '''RAREMETALWORKER''' or [http://genome.sph.umich.edu/wiki/Rvtests '''rvtests'''], a file with list of studies to be included and a group file if gene-level meta-analysis is expected. | |
| − | |||
| − | |||
| − | |||
| − | ===== | + | =====Summary Statistics===== |
| − | + | Files containing summary statistics and LD matrices generated by '''RAREMETALWORKER''' should be compressed and [http://samtools.sourceforge.net/tabix.shtml '''tabix'''] indexed using the following commands (Note in RAREMETALWORKER, if --zip is specified, these .gz and .tbi files will be automatically generated): | |
| − | |||
| − | |||
| − | |||
| − | + | bgzip study1.singlevar.score.txt | |
| + | tabix -s 1 -b 2 -e 2 -c "#" study1.singlevar.score.txt.gz | ||
| + | bgzip study1.singlevar.cov.txt | ||
| + | tabix -s 1 -b 2 -e 2 -c "#" study1.singlevar.cov.txt.gz | ||
| − | + | Files containing summary statistics and LD matrices generated by '''rvtests''' should be compressed and [http://samtools.sourceforge.net/tabix.shtml '''tabix'''] indexed using the following commands: | |
| − | |||
| − | + | bgzip study1.MetaScore.assoc | |
| − | + | tabix -s 1 -b 2 -e 2 -S 1 study1.MetaScore.assoc.gz | |
| − | + | tabix -s 1 -b 2 -e 2 -S 1 study1.MetaCov.assoc.gz | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==== List of Studies ==== | + | =====List of Studies===== |
| − | * -- | + | * --summaryFiles option is crucial for '''RAREMETAL''' to work. Ignoring this option would lead to FATAL ERROR and '''RAREMETAL''' would stop. |
* The file should contain the path and prefix of the studies you want to include. | * The file should contain the path and prefix of the studies you want to include. | ||
| − | * If there is one or more studies that you want to excluded from your list, but want to save some effort of generating a new file, you can put a "#" in front of the line of record. ''' | + | * If there is one or more studies that you want to excluded from your list, but want to save some effort of generating a new file, you can put a "#" in front of the line of record. '''RAREMETAL''' would automatically exclude that study from meta analysis. An example list of summary file is in the following: |
| − | + | ||
| + | /net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/TwinsUK/TwinsUK.TG.singlevar.score.txt.gz | ||
| + | #/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/HUNT/RareMetalWorker/HUNT_MI_case.TG.singlevar.score.txt.gz | ||
| − | + | * When gene-level analysis is requested, --covFiles option should be used to specify the covariance files. An example file is: | |
| − | |||
| − | + | /net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/TwinsUK/TwinsUK.TG.singlevar.cov.txt.gz | |
| + | #/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/HUNT/RareMetalWorker/HUNT_MI_case.TG.singlevar.cov.txt.gz | ||
| − | + | * The above example study name file guides '''RAREMETAL''' to look for summary statistics from TwinsUK study only, because "HUNT" study is commented out. The following two files are needed for '''RAREMETAL''' to perform further analysis together with their tabix index file are needed. | |
| − | |||
| − | + | * Please sepcify --dosage option if input files were generated from dosage instead of genotype. | |
| − | ===== | + | =====Group Rare Variants===== |
| + | |||
| + | ====== From a Group File ====== | ||
* Grouping methods are only necessary when doing gene-based or group-based burden tests in meta-analysis. | * Grouping methods are only necessary when doing gene-based or group-based burden tests in meta-analysis. | ||
* If none of the grouping method is specified, then only single variant meta-analysis will be performed. | * If none of the grouping method is specified, then only single variant meta-analysis will be performed. | ||
| Line 92: | Line 94: | ||
* MARKER_ID must be in the following format: | * MARKER_ID must be in the following format: | ||
CHR:POS:REF:ALT | CHR:POS:REF:ALT | ||
| − | * | + | * An example group file is: |
PLEKHN1 1:901922:G:A 1:901923:C:A 1:902088:G:A 1:902128:C:T 1:902133:C:G 1:902176:C:T 1:905669:C:G | PLEKHN1 1:901922:G:A 1:901923:C:A 1:902088:G:A 1:902128:C:T 1:902133:C:G 1:902176:C:T 1:905669:C:G | ||
HES4 1:934735:A:C 1:934770:G:A 1:934801:C:T 1:935085:G:A 1:935089:C:G | HES4 1:934735:A:C 1:934770:G:A 1:934801:C:T 1:935085:G:A 1:935089:C:G | ||
| Line 99: | Line 101: | ||
C1orf159 1:1021285:G:T 1:1021302:T:C 1:1021315:A:C 1:1021386:G:A 1:1022534:C:T 1:1025751:C:T 1:1026913:C:T | C1orf159 1:1021285:G:T 1:1021302:T:C 1:1021315:A:C 1:1021386:G:A 1:1022534:C:T 1:1025751:C:T 1:1026913:C:T | ||
| − | ===== | + | ====== From an Annotated VCF File ====== |
| − | + | If --groupFile option is '''NOT''' specified, '''RAREMETAL''' will look for an annotated vcf file as blue print for variants to group. Users are also allowed to generate a vcf file based on the superset of variants from pooled samples, and annotate outside RAREMETAL. Then, annotated vcf file can be used as input for RAREMETAL for gene-level meta-analysis, or group files can be generated based on the annotated vcf file. Detailed description of these options are [[Rare-Metal#Group_Rare_Variants_from_Annotated_VCF|'''available''']]. There are also [[Rare-Metal#Example_Command_lines|'''examples''']] of this usage at the bottom of this page. | |
| + | |||
| + | ==== QC Options ==== | ||
| + | * '''RAREMETAL''' allows filtering of variants from individual studies by their HWE pvalue and call rate, which are generated as part of the output from '''RAREMETALWORKER''' or [http://genome.sph.umich.edu/wiki/Rvtests '''rvtests''']. | ||
| + | * To filter by HWE p-values, --hwe option should be used. The default is 0.0, which means not filtering any of the variants. | ||
| + | * To filter by call rate, --callRate option can be specified. The default is 0.0, which allows no filtering utilized. | ||
| + | |||
| + | ==== Association Options==== | ||
| + | * Currently, CMC type burden test, Madsen-Browning burden test, Variable Threshold burden test and SKAT are provided in '''RAREMETAL''', by specifying --burden, --MB, --VT and --SKAT. | ||
| + | * --maf specifies the minor allele frequency cutoff when doing gene-based or group-based burden tests. Variants with maf '''above''' this threshold will be ignored. The default is maf<0.05. | ||
| + | * In '''a single study''' of sample size N, if a site is monomorphic or not reported in vcf/ped, it is considered that the sample size of this study is not large enough to sample the rare allele. Thus, this study contributes 2*N reference alleles and 0 alternative allele towards meta-analysis. To let such studies contribute no alleles towards pooled allele frequency, specify --altMAF. | ||
| + | |||
| + | ==== Conditional Analysis==== | ||
| + | * To decide whether a signal is caused by shadowing a significant common variant nearby, '''RAREMETAL''' also enables conditional analysis with a list of variants to be conditioned upon provided in a file as input for --condition option. An example input file should be space or tab delimited as in the following. When alleles do not match the ref and alt alleles from samples, the variant will be skipped from conditional analysis. | ||
| + | |||
| + | 1:861349:C:T 1:905901:G:A 20:986998:G:C 22:3670691:A:G | ||
| + | |||
| + | == Additional Analysis Options == | ||
| + | |||
| + | === Generate a VCF File to Annotate Outside RAREMETAL === | ||
| + | * --writeVCF allows user to write a VCF file including pooled single variants from all studies. Then users can use their favorite annotation tool to annotate the VCF file. After annotating the VCF file, users can use that file as input for '''RAREMETAL''' for further gene-based or region-based meta analysis. | ||
| + | * The output vcf file will be name as: yourPrefix.pooled.variants.vcf. An example output vcf file is in the following: | ||
| + | #CHROM POS ID REF ALT QUAL FILTER INFO | ||
| + | 1 115658497 115658497 G A . . ALT_AF=0.380906; | ||
| + | 2 74688884 74688884 G A . . ALT_AF=8.33611e-05; | ||
| + | 3 121414217 121414217 C A . . ALT_AF=0.0747833; | ||
| + | ===Annotation=== | ||
| + | * RAREMETAL automatically recognizes the annotation format generated by [[TabAnno | '''ANNO''']] or [[EPACTS#Annotating_VCF_file_using_EPACTS | '''EPACTS''']]. | ||
| + | * To annotate a the VCF generated in previous step, you can use the following command: | ||
| + | ./anno --in your.in.vcf.gz --out your.out.vcf.gz | ||
| + | |||
| + | === Group Rare Variants from Annotated VCF === | ||
| + | * If --groupFile option is '''NOT''' specified, '''RAREMETAL''' will look for an annotated vcf file as blue print for variants to group. | ||
* The annotated VCF file should be specified using --annotatedVcf option. | * The annotated VCF file should be specified using --annotatedVcf option. | ||
* --annotation should be used with --annotatedVcf together when specific category of functional variants are of interest to be grouped. For example, if grouping nonsynonymous and splicing variants are of interests, the following should be included in command line: | * --annotation should be used with --annotatedVcf together when specific category of functional variants are of interest to be grouped. For example, if grouping nonsynonymous and splicing variants are of interests, the following should be included in command line: | ||
| + | * (! only available after v4.13.8) when --annotation is not specified, raremetal groups all non-intergenic variants. | ||
--annotatedVcf your.annotated.vcf --annotation nonsyn/splicing | --annotatedVcf your.annotated.vcf --annotation nonsyn/splicing | ||
| Line 117: | Line 152: | ||
* Notice that each variant is allowed to have more than one annotations; but each annotation should start with a new key "ANNO=" followed by annotation:genename:other transcript information. | * Notice that each variant is allowed to have more than one annotations; but each annotation should start with a new key "ANNO=" followed by annotation:genename:other transcript information. | ||
| + | * Generated group file will be named test.groupfile under your running directory. | ||
| − | === | + | ===Options for Report Generation=== |
| − | + | * --correctGC generates QQ plots and manhattan plots with pvalues corrected using genomic control. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
* --prefix allows customized prefix for output files. | * --prefix allows customized prefix for output files. | ||
| − | |||
* --longOutput allows users to output not only burden test results but also the single variant results (allele frequencies, effect sizes, and p-values) for the variants being grouped together. Please refer to the output files section for detailed explanation and examples. | * --longOutput allows users to output not only burden test results but also the single variant results (allele frequencies, effect sizes, and p-values) for the variants being grouped together. Please refer to the output files section for detailed explanation and examples. | ||
| − | * --tabulateHits works with --hitsCutoff together to generate reports for genes that have p-value less than specified cutoff from burden tests or SKAT. The default cutoff of p-value for genes to be reported is 1.0e-06, which can be specified by --hitsCutoff option. For more explanations and examples, please go to [[ | + | * --tabulateHits works with --hitsCutoff together to generate reports for genes that have p-value less than specified cutoff from burden tests or SKAT. The default cutoff of p-value for genes to be reported is 1.0e-06, which can be specified by --hitsCutoff option. For more explanations and examples, please go to [[Rare-Metal#TABULATED_HITS| Tabulated Hits]]. |
| + | |||
| + | ===Miscellaneous Options=== | ||
| + | * --tabix allows rapid analysis when number of groups/genes of interests are small. Currently, when number of groups is less than 100, --tabix option is automatically turned on. | ||
| − | === Output | + | == Reports Generated by RAREMETAL == |
| + | === Single Variant Meta Analysis Output === | ||
| − | ==== | + | ==== TABLES ==== |
| − | * Single variant meta analysis output has the following components: header and | + | * Single variant meta analysis output has the following components: header, results and footnote. |
* Header lines start with "##" shows summary of the meta analysis including method used, number of studies, and total sample size. | * Header lines start with "##" shows summary of the meta analysis including method used, number of studies, and total sample size. | ||
* Header line starts with "#" are column headers for results table. | * Header line starts with "#" are column headers for results table. | ||
| + | * Footnote also starts with "#", where genomic controls from each study and the overall sample are reported. | ||
* An example single variant meta analysis output is shown below: | * An example single variant meta analysis output is shown below: | ||
| Line 166: | Line 191: | ||
EFFECT_SIZE: Alternative Allele Effect Size | EFFECT_SIZE: Alternative Allele Effect Size | ||
DIRECTION_BY_STUDY: Effect size direction of alternative allele from each study. | DIRECTION_BY_STUDY: Effect size direction of alternative allele from each study. | ||
| − | The order of study is consistent with the order of studies listed in the input file for option -- | + | The order of study is consistent with the order of studies listed in the input file for option --summaryFiles. |
"?" means the variant is not observed or monomorphic from the study. | "?" means the variant is not observed or monomorphic from the study. | ||
| − | "!" means the variant observed from this study has different alleles from those | + | "!" means the variant observed from this study has different alleles from those in the first study. |
| + | |||
| + | ==== PLOTS==== | ||
| − | + | '''RAREMETAL''' generates QQ plots and manhattan plots from single variant meta-analysis by default. Three QQ plots are generated, one with all variants included, one of variants with maf<0.05 and one of variants with maf<0.01. All plots are saved in a pdf file named yourPrefix.meta.plots.pdf. Genomic controls are also reported in the title of plots. When --correctGC option is specified, GC corrected plots are also generated. | |
| − | + | {| border="1" cellpadding="5" cellspacing="0" align="center" | |
| + | |- | ||
| + | | align="center" width="100" | [[File:QQ.png]] | ||
| + | |- | ||
| + | | align="center" width="200" | [[File:Single_var_manhattan.png]] | ||
| + | |} | ||
| − | ===== | + | === Gene-level Tests Meta-Analysis Output === |
| − | + | ||
| + | |||
| + | ==== LONG TABLES ==== | ||
| + | When --longOutput is used, output includes both burden test results of genes and single variant results of the variants included in burden tests. Here is an example of output file from SKAT when --longOutput is specified. | ||
##Method=Burden | ##Method=Burden | ||
##STUDY_NUM=2 | ##STUDY_NUM=2 | ||
| Line 182: | Line 217: | ||
KLHL17 2 1:897285:A:G;1:898869:C:T 0.0148408,0.00108369 -0.0502034,-0.0256403 0.528269,0.934606 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878 | KLHL17 2 1:897285:A:G;1:898869:C:T 0.0148408,0.00108369 -0.0502034,-0.0256403 0.528269,0.934606 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878 | ||
| − | ==== | + | ==== SHORT TABLES ==== |
| − | + | Otherwise, single variant results of variants included in burden tests will not be included in the output. Here is an example of output file from SKAT when --longOutput is not specified. | |
##Method=Burden | ##Method=Burden | ||
| Line 192: | Line 227: | ||
KLHL17 2 1:897285:A:G;1:898869:C:T 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878 | KLHL17 2 1:897285:A:G;1:898869:C:T 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878 | ||
| − | ==== | + | ==== TABULATED HITS ==== |
* When --tabulateHits is specified, top hits from Burden tests will be generated. Each method will have an individual tabulated file generated. The purpose of this tabulated file is to list burden test results of top hits together with single variant results from variants being grouped in burden tests. The difference between this file and the standard long-format output file from burden test is that each row of the file represents a single variant that is included in the gene for burden test. This format allows each sorting on users end. | * When --tabulateHits is specified, top hits from Burden tests will be generated. Each method will have an individual tabulated file generated. The purpose of this tabulated file is to list burden test results of top hits together with single variant results from variants being grouped in burden tests. The difference between this file and the standard long-format output file from burden test is that each row of the file represents a single variant that is included in the gene for burden test. This format allows each sorting on users end. | ||
| − | * | + | * Tabulated top hits are saved in the file: |
yourPrefix.meta.tophits.youMethod.tbl (example files names: TG.meta.tophits.burden.tbl, LDL.meta.tophits.SKAT.tbl) | yourPrefix.meta.tophits.youMethod.tbl (example files names: TG.meta.tophits.burden.tbl, LDL.meta.tophits.SKAT.tbl) | ||
| Line 220: | Line 255: | ||
* According to the example above, PCSK9 had a p-value of 7.54587e-11 from the gene-based burden test, where three variants from this gene were included. Another hit from this meta analysis is APOE, where only one variant was included in the burden test. | * According to the example above, PCSK9 had a p-value of 7.54587e-11 from the gene-based burden test, where three variants from this gene were included. Another hit from this meta analysis is APOE, where only one variant was included in the burden test. | ||
| − | ==== | + | ==== PLOTS ==== |
| + | '''RAREMETAL''' generates QQ plots and manhattan plots from single variant and gene-level meta-analysis by default. Example QQ plots and manhattan plots are: | ||
| + | {| border="1" cellpadding="5" cellspacing="0" align="center" | ||
| + | |- | ||
| + | | align="center" width="200" | [[File:manhattan.png]] | ||
| + | |} | ||
| − | * A log file is automatically generated by ''' | + | ==== LOG ==== |
| + | |||
| + | * A log file is automatically generated by '''RAREMETAL''' to save the parameters in effect. An example is in the following: | ||
The following parameters are in effect: | The following parameters are in effect: | ||
| Line 232: | Line 274: | ||
Grouping Methods: | Grouping Methods: | ||
============================ | ============================ | ||
| − | --groupFile [] | + | --groupFile [genes.file] |
| − | --annotatedVcf [ | + | --annotatedVcf [] |
--annotation [] | --annotation [] | ||
--writeVcf [OFF] | --writeVcf [OFF] | ||
| Line 248: | Line 290: | ||
--SKAT [false] | --SKAT [false] | ||
--VT [false] | --VT [false] | ||
| + | --condition [condition.file] | ||
Other Options: | Other Options: | ||
============================ | ============================ | ||
| + | --tabix [OFF] | ||
| + | --correctGC [ON] | ||
--prefix [test] | --prefix [test] | ||
--maf [0.05] | --maf [0.05] | ||
| Line 256: | Line 301: | ||
--tabulateHits [false] | --tabulateHits [false] | ||
--hitsCutoff [1e-06] | --hitsCutoff [1e-06] | ||
| + | --dosage [false] | ||
| + | --altMAF [false] | ||
| − | ==Example | + | ==Example Command lines== |
* Here is an example command line to do single variant meta analysis only: | * Here is an example command line to do single variant meta analysis only: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --prefix yourPrefix |
* When you want to do all burden tests using a group file to specify which variants to group: | * When you want to do all burden tests using a group file to specify which variants to group: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --burden --MB --SKAT --VT --maf 0.01 --prefix yourPrefix |
(NOTE: this will generate single variant meta analysis result and the short format output for burden test results.) | (NOTE: this will generate single variant meta analysis result and the short format output for burden test results.) | ||
* Here is how to do all SKAT meta analysis using a group file and request a long format output together with tabulated hits: | * Here is how to do all SKAT meta analysis using a group file and request a long format output together with tabulated hits: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --prefix yourPrefix |
* Here is an example of adding QC filters to variants when doing meta analysis. | * Here is an example of adding QC filters to variants when doing meta analysis. | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --hwe 1e-06 --callRate 0.98 --prefix yourPrefix |
* Here is how to do the same thing but reading grouping information from an annotated VCF file: | * Here is how to do the same thing but reading grouping information from an annotated VCF file: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --annotatedVcf your.annotated.vcf --annotation nonsyn/stop/splicing --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --hwe 1e-06 --callRate 0.98 --prefix yourPrefix |
| − | * If you want to write a VCF file of pooled variants from all studies, annotate them using your favorite annotation program, and then come back to ''' | + | * If you want to write a VCF file of pooled variants from all studies, annotate them using your favorite annotation program, and then come back to '''RAREMETAL''' with the annotate VCF file to do burden tests: |
First, use the following command to write the VCF file: | First, use the following command to write the VCF file: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --writeVcf --prefix yourPrefix |
| − | Second, annotate the VCF file using your favorite annotation program. (Annotated VCF file has to follow the format described here: [[ | + | Second, annotate the VCF file using your favorite annotation program. (Annotated VCF file has to follow the format described here: [[Rare-Metal#Group_Rare_Variants_from_Annotated_VCF|annotated VCF format]]) |
Third, use the following command to do meta analysis: | Third, use the following command to do meta analysis: | ||
| − | ./raremetal -- | + | ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --annotatedVcf your.annotated.vcf --annotation nonsyn/splicing/stop --burden --MB --SKAT --VT --maf 0.01 --prefix yourPrefix |
| + | |||
| + | ==Other Useful Info== | ||
| + | |||
| + | * Summary specs can be found [[Summary Files Specification for RAREMETAL]] | ||
==TUTORIAL== | ==TUTORIAL== | ||
| − | * For a comprehensive tutorial of | + | * For a comprehensive tutorial of RAREMETALWORKER and RAREMETAL using example data sets, please go to the following: |
| + | |||
| + | [http://genome.sph.umich.edu/wiki/Tutorial:_RareMETAL '''RAREMETAL Tutorial'''] | ||
| + | |||
| + | * For a brief tutorial of rvtests, please go to: | ||
| − | [http://genome.sph.umich.edu/wiki/ | + | [http://genome.sph.umich.edu/wiki/Rvtests '''rvtests'''] |
| − | == | + | ==CONTACT== |
| + | Please email Andy Boughton (abought at umich dot edu) for questions. | ||
| − | + | Also check [[Raremetal Incoming updates | '''Known issues and incoming update in next version''']] to see if your problem has been reported before | |
| − | |||
| − | |||
| − | |||
Latest revision as of 13:22, 20 May 2019
Useful Wiki Pages
- Git hub page: https://github.com/statgen/Raremetal
- The Change Log
- The DOWNLOAD page
- The RAREMETAL FAQ
The rvtests tool for rare-variant association analysis can also generate output compatible with RAREMETAL.
Brief Description
RAREMETAL is a computationally efficient tool for meta-analysis of rare variants using sequencing or genotyping array data. It takes summary statistics and LD matrices generated by RAREMETALWORKER or rvtests, handles related and unrelated individuals, and supports both single variant and burden meta-analysis. It generates high quality plots by default and has options that allow users to build reports at different levels.
RAREMETAL is developed by Shuang Feng, Dajiang Liu and Gonçalo Abecasis. A R-package written by Dajiang Liu using the same methodology is available.
Key Features
RAREMETAL has the following features:
- Performs gene-based or region-based meta analysis using Burden tests with the following methods: CMC_counts, Madsen-Browning, SKAT, and Variable Threshold.
- Performs single variant metal-analysis by default.
- Allows customized groups of variants to be tested.
- Allows conditional analysis to be performed in both gene-level meta-analysis and single variants meta-analysis.
- Generate QQ plots and manhattan plots by default.
Approach
The key idea behind meta-analysis with RAREMETAL is that various gene-level test statistics can be reconstructed from single variant score statistics and that, when the linkage disequilibrium relationships between variants are known, the distribution of these gene-level statistics can be derived and used to evaluate signifi-cance. Single variant statistics are calculated using the Cochran-Mantel-Haenszel method. Our method has been published in Liu et. al in Nature Genetics. Please go to method for details.
Download and Installation
We have tested compilation using our source code on several platforms including Linux, and Mac OS X.
For source code and executables together with instructions of building from source, please go to DOWNLOAD source and executables.
For questions about compilation, please go to FAQ.
Basic Usage Instructions
RAREMETAL is a command line tool. It is typically run from a Linux or Unix prompt by invoking the command raremetal. In the following are descriptions of basic usage for meta analysis. A detailed TUTORIAL with toy data are also available.
Prepare Input Files
RAREMETAL requires the following basic input files: summary statistics and covariance matrices of score statistics generated by RAREMETALWORKER or rvtests, a file with list of studies to be included and a group file if gene-level meta-analysis is expected.
Summary Statistics
Files containing summary statistics and LD matrices generated by RAREMETALWORKER should be compressed and tabix indexed using the following commands (Note in RAREMETALWORKER, if --zip is specified, these .gz and .tbi files will be automatically generated):
bgzip study1.singlevar.score.txt tabix -s 1 -b 2 -e 2 -c "#" study1.singlevar.score.txt.gz bgzip study1.singlevar.cov.txt tabix -s 1 -b 2 -e 2 -c "#" study1.singlevar.cov.txt.gz
Files containing summary statistics and LD matrices generated by rvtests should be compressed and tabix indexed using the following commands:
bgzip study1.MetaScore.assoc tabix -s 1 -b 2 -e 2 -S 1 study1.MetaScore.assoc.gz tabix -s 1 -b 2 -e 2 -S 1 study1.MetaCov.assoc.gz
List of Studies
- --summaryFiles option is crucial for RAREMETAL to work. Ignoring this option would lead to FATAL ERROR and RAREMETAL would stop.
- The file should contain the path and prefix of the studies you want to include.
- If there is one or more studies that you want to excluded from your list, but want to save some effort of generating a new file, you can put a "#" in front of the line of record. RAREMETAL would automatically exclude that study from meta analysis. An example list of summary file is in the following:
/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/TwinsUK/TwinsUK.TG.singlevar.score.txt.gz #/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/HUNT/RareMetalWorker/HUNT_MI_case.TG.singlevar.score.txt.gz
- When gene-level analysis is requested, --covFiles option should be used to specify the covariance files. An example file is:
/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/TwinsUK/TwinsUK.TG.singlevar.cov.txt.gz #/net/fantasia/home/sfengsph/prj/raremetal/raremetal/bin/META/HUNT/RareMetalWorker/HUNT_MI_case.TG.singlevar.cov.txt.gz
- The above example study name file guides RAREMETAL to look for summary statistics from TwinsUK study only, because "HUNT" study is commented out. The following two files are needed for RAREMETAL to perform further analysis together with their tabix index file are needed.
- Please sepcify --dosage option if input files were generated from dosage instead of genotype.
Group Rare Variants
From a Group File
- Grouping methods are only necessary when doing gene-based or group-based burden tests in meta-analysis.
- If none of the grouping method is specified, then only single variant meta-analysis will be performed.
- With --groupFile option, you can specify particular set of variants to be grouped for burden tests.
- The group file must be a tab or space delimited file in the following format:
GROUP_ID MARKER1_ID MARKER2_ID MARKER3_ID ...
- MARKER_ID must be in the following format:
CHR:POS:REF:ALT
- An example group file is:
PLEKHN1 1:901922:G:A 1:901923:C:A 1:902088:G:A 1:902128:C:T 1:902133:C:G 1:902176:C:T 1:905669:C:G HES4 1:934735:A:C 1:934770:G:A 1:934801:C:T 1:935085:G:A 1:935089:C:G ISG15 1:949422:G:A 1:949491:G:A 1:949502:C:T 1:949608:G:A 1:949802:G:A 1:949832:G:A AGRN 1:970687:C:T 1:976963:A:G 1:977028:G:T 1:977356:C:T 1:977396:G:A 1:978628:C:T 1:978645:G:A C1orf159 1:1021285:G:T 1:1021302:T:C 1:1021315:A:C 1:1021386:G:A 1:1022534:C:T 1:1025751:C:T 1:1026913:C:T
From an Annotated VCF File
If --groupFile option is NOT specified, RAREMETAL will look for an annotated vcf file as blue print for variants to group. Users are also allowed to generate a vcf file based on the superset of variants from pooled samples, and annotate outside RAREMETAL. Then, annotated vcf file can be used as input for RAREMETAL for gene-level meta-analysis, or group files can be generated based on the annotated vcf file. Detailed description of these options are available. There are also examples of this usage at the bottom of this page.
QC Options
- RAREMETAL allows filtering of variants from individual studies by their HWE pvalue and call rate, which are generated as part of the output from RAREMETALWORKER or rvtests.
- To filter by HWE p-values, --hwe option should be used. The default is 0.0, which means not filtering any of the variants.
- To filter by call rate, --callRate option can be specified. The default is 0.0, which allows no filtering utilized.
Association Options
- Currently, CMC type burden test, Madsen-Browning burden test, Variable Threshold burden test and SKAT are provided in RAREMETAL, by specifying --burden, --MB, --VT and --SKAT.
- --maf specifies the minor allele frequency cutoff when doing gene-based or group-based burden tests. Variants with maf above this threshold will be ignored. The default is maf<0.05.
- In a single study of sample size N, if a site is monomorphic or not reported in vcf/ped, it is considered that the sample size of this study is not large enough to sample the rare allele. Thus, this study contributes 2*N reference alleles and 0 alternative allele towards meta-analysis. To let such studies contribute no alleles towards pooled allele frequency, specify --altMAF.
Conditional Analysis
- To decide whether a signal is caused by shadowing a significant common variant nearby, RAREMETAL also enables conditional analysis with a list of variants to be conditioned upon provided in a file as input for --condition option. An example input file should be space or tab delimited as in the following. When alleles do not match the ref and alt alleles from samples, the variant will be skipped from conditional analysis.
1:861349:C:T 1:905901:G:A 20:986998:G:C 22:3670691:A:G
Additional Analysis Options
Generate a VCF File to Annotate Outside RAREMETAL
- --writeVCF allows user to write a VCF file including pooled single variants from all studies. Then users can use their favorite annotation tool to annotate the VCF file. After annotating the VCF file, users can use that file as input for RAREMETAL for further gene-based or region-based meta analysis.
- The output vcf file will be name as: yourPrefix.pooled.variants.vcf. An example output vcf file is in the following:
#CHROM POS ID REF ALT QUAL FILTER INFO 1 115658497 115658497 G A . . ALT_AF=0.380906; 2 74688884 74688884 G A . . ALT_AF=8.33611e-05; 3 121414217 121414217 C A . . ALT_AF=0.0747833;
Annotation
- RAREMETAL automatically recognizes the annotation format generated by ANNO or EPACTS.
- To annotate a the VCF generated in previous step, you can use the following command:
./anno --in your.in.vcf.gz --out your.out.vcf.gz
Group Rare Variants from Annotated VCF
- If --groupFile option is NOT specified, RAREMETAL will look for an annotated vcf file as blue print for variants to group.
- The annotated VCF file should be specified using --annotatedVcf option.
- --annotation should be used with --annotatedVcf together when specific category of functional variants are of interest to be grouped. For example, if grouping nonsynonymous and splicing variants are of interests, the following should be included in command line:
- (! only available after v4.13.8) when --annotation is not specified, raremetal groups all non-intergenic variants.
--annotatedVcf your.annotated.vcf --annotation nonsyn/splicing Note: this allows you to group variants that are annotated starting with nonsyn or splicing (not case-sensitive).
- Special format for the annotated VCF file is required: all annotation information should be coded in INFO field in VCF file, starting with the key "ANNO=". An example annotated VCF file is in the following:
#CHROM POS ID REF ALT QUAL FILTER INFO 1 19208194 . G A 100 PASS AC=3;ANNO=nonsynonymous:ALDH4A1:NM_170726:exon8:c.C866T:p.P289L,ALDH4A1:NM_001161504:exon8:c.C686T:p.P229L,ALDH4A1:NM_003748:exon8:c.C866T:p.P289L,; ANNO=splicing:ALDH4A1 1 19208293 . G C 100 PASS AC=7;STUDIES=5;MAC=7;MAF=0.001;DESIGN=TBD_ASSAY;DSCORE=1.00; ANNO=nonsynonymous:ALDH4A1:NM_170726:exon8:c.C767G:p.P256R,ALDH4A1:NM_001161504:exon8:c.C587G:p.P196R,ALDH4A1:NM_003748:exon8:c.C767G:p.P256R,
- Notice that each variant is allowed to have more than one annotations; but each annotation should start with a new key "ANNO=" followed by annotation:genename:other transcript information.
- Generated group file will be named test.groupfile under your running directory.
Options for Report Generation
- --correctGC generates QQ plots and manhattan plots with pvalues corrected using genomic control.
- --prefix allows customized prefix for output files.
- --longOutput allows users to output not only burden test results but also the single variant results (allele frequencies, effect sizes, and p-values) for the variants being grouped together. Please refer to the output files section for detailed explanation and examples.
- --tabulateHits works with --hitsCutoff together to generate reports for genes that have p-value less than specified cutoff from burden tests or SKAT. The default cutoff of p-value for genes to be reported is 1.0e-06, which can be specified by --hitsCutoff option. For more explanations and examples, please go to Tabulated Hits.
Miscellaneous Options
- --tabix allows rapid analysis when number of groups/genes of interests are small. Currently, when number of groups is less than 100, --tabix option is automatically turned on.
Reports Generated by RAREMETAL
Single Variant Meta Analysis Output
TABLES
- Single variant meta analysis output has the following components: header, results and footnote.
- Header lines start with "##" shows summary of the meta analysis including method used, number of studies, and total sample size.
- Header line starts with "#" are column headers for results table.
- Footnote also starts with "#", where genomic controls from each study and the overall sample are reported.
- An example single variant meta analysis output is shown below:
##Method=SinglevarScore ##STUDY_NUM=2 ##TotalSampleSize=14308 #CHROM POS REF ALT POOLED_ALT_AF EFFECT_SIZE DIRECTION_BY_STUDY PVALUE 1 115658497 G A 0.380906 0.00954332 ++ 0.45828 2 74688884 G A 8.33611e-05 -0.196387 -! 0.845372 3 121414217 C A 0.0747833 0.0216982 -+ 0.34453 6 137245814 G C 0.000803746 0.105693 ++ 0.601805
- A detailed explanation of each column is in the following:
CHROM: Chromosome Name
POS: Variant Position
REF: Reference Allele Label
ALT: Alternative Allele Label
POOLED_ALT_AF: Pooled Alternative Allele Frequency
EFFECT_SIZE: Alternative Allele Effect Size
DIRECTION_BY_STUDY: Effect size direction of alternative allele from each study.
The order of study is consistent with the order of studies listed in the input file for option --summaryFiles.
"?" means the variant is not observed or monomorphic from the study.
"!" means the variant observed from this study has different alleles from those in the first study.
PLOTS
RAREMETAL generates QQ plots and manhattan plots from single variant meta-analysis by default. Three QQ plots are generated, one with all variants included, one of variants with maf<0.05 and one of variants with maf<0.01. All plots are saved in a pdf file named yourPrefix.meta.plots.pdf. Genomic controls are also reported in the title of plots. When --correctGC option is specified, GC corrected plots are also generated.
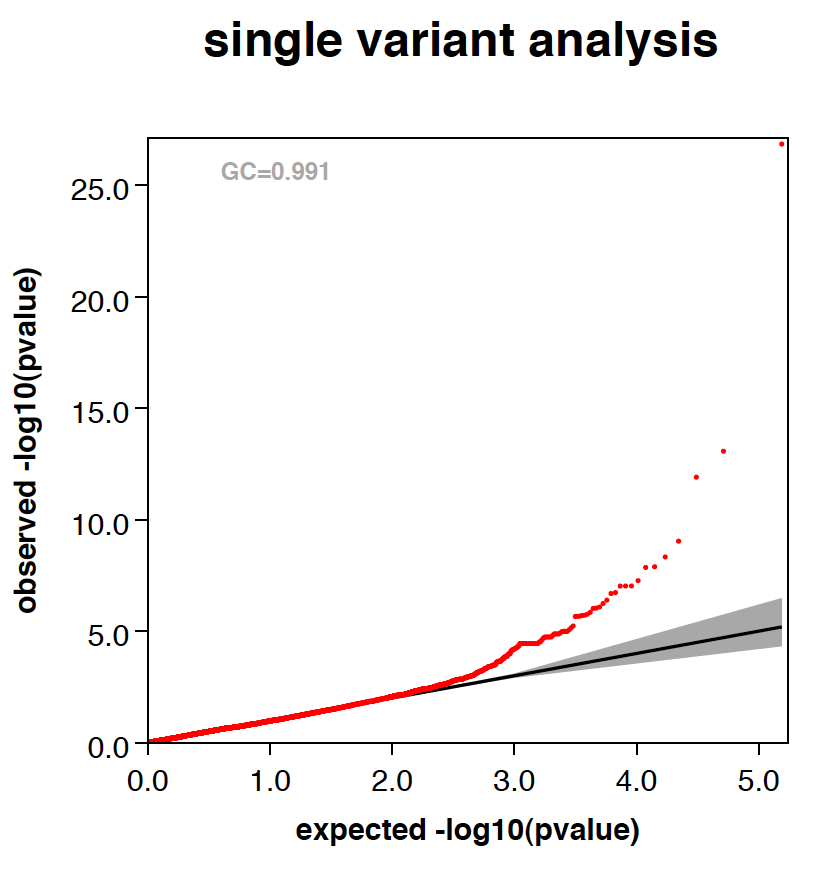
|
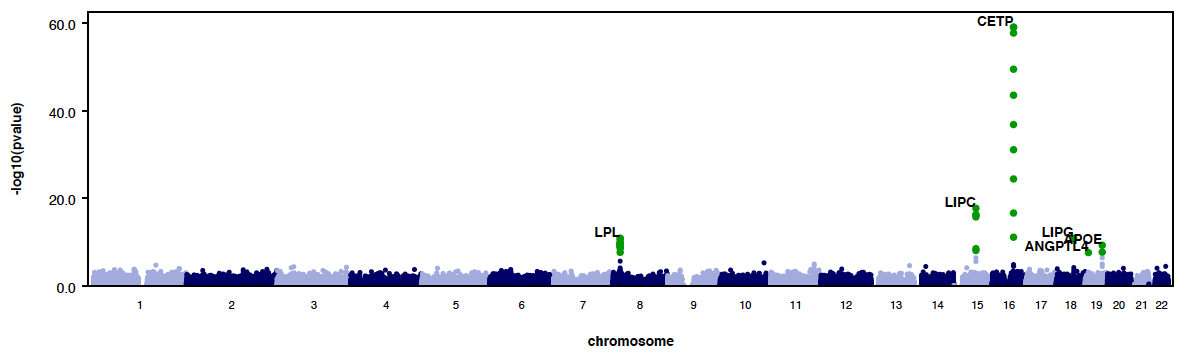
|
Gene-level Tests Meta-Analysis Output
LONG TABLES
When --longOutput is used, output includes both burden test results of genes and single variant results of the variants included in burden tests. Here is an example of output file from SKAT when --longOutput is specified.
##Method=Burden ##STUDY_NUM=2 ##TotalSampleSize=14308 #GROUPNAME NUM_VAR VARs MAFs SINGLEVAR_EFFECTs SINGLEVAR_PVALUEs AVG_AF MIN_AF MAX_AF EFFECT_SIZE PVALUE NOC2L 7 1:880502:C:T;1:881918:G:A;1:887799:C:T;1:888659:T:C;1:889238:G:A;1:891591:C:T;1:892380:G:A 0.000166722,0.0242172,0.0109203,0.0355845,0.0333729,0.00700233,0.00200067 -0.183575,-0.00228307,-0.0598337,0.0220595,0.0229464,-0.0302768,-0.0200417 0.790161,0.953446,0.515806,0.503548,0.499251,0.791773,0.926625 0.0161807 0.000166722 0.0355845 0.00667875 0.662531 KLHL17 2 1:897285:A:G;1:898869:C:T 0.0148408,0.00108369 -0.0502034,-0.0256403 0.528269,0.934606 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878
SHORT TABLES
Otherwise, single variant results of variants included in burden tests will not be included in the output. Here is an example of output file from SKAT when --longOutput is not specified.
##Method=Burden ##STUDY_NUM=2 ##TotalSampleSize=14308 #GROUPNAME NUM_VAR VARs AVG_AF MIN_AF MAX_AF EFFECT_SIZE PVALUE NOC2L 7 1:880502:C:T;1:881918:G:A;1:887799:C:T;1:888659:T:C;1:889238:G:A;1:891591:C:T;1:892380:G:A 0.0161807 0.000166722 0.0355845 0.00667875 0.662531 KLHL17 2 1:897285:A:G;1:898869:C:T 0.00796222 0.00108369 0.0148408 -0.0484494 0.528878
TABULATED HITS
- When --tabulateHits is specified, top hits from Burden tests will be generated. Each method will have an individual tabulated file generated. The purpose of this tabulated file is to list burden test results of top hits together with single variant results from variants being grouped in burden tests. The difference between this file and the standard long-format output file from burden test is that each row of the file represents a single variant that is included in the gene for burden test. This format allows each sorting on users end.
- Tabulated top hits are saved in the file:
yourPrefix.meta.tophits.youMethod.tbl (example files names: TG.meta.tophits.burden.tbl, LDL.meta.tophits.SKAT.tbl)
- The following items are tabulated in the output:
GENE: Gene name.
METHOD: Burden test used.
GENE_PVALUE: P-value from gene-based burden tests.
MAF_CUTOFF: MAF cutoff used when doing gene-based tests.
ACTUAL_CUTOFF: Actual MAF cutoff used. (This will be different from MAF_CUTOFF only for Variable Threshold method.
Otherwise, it will be the same as MAF_CUTOFF.)
VAR: Variant name in CHR:POS:REF:ALT format.
MAF: Single variant pooled MAF from all samples.
EFFSIZE: Effect size from single variant meta analysis.
PVALUE: Pvalue from single variant meta analysis.
- An example of tabulated hits from a standard burden test with maf<0.05 as criterion is shown in the following:
GENE METHOD GENE_PVALUE MAF_CUTOFF ACTUAL_CUTOFF VARS MAFS EFFSIZES PVALUES PCSK9 BURDEN_0.050 7.54587e-11 0.05 0.05 1:55505647:G:T 0.0396631 -0.442192 2.10159e-46 PCSK9 BURDEN_0.050 7.54587e-11 0.05 0.05 1:55518371:G:A 0.0237138 0.0548733 0.430246 PCSK9 BURDEN_0.050 7.54587e-11 0.05 0.05 1:55529187:G:A 0.0433324 0.0946321 0.00129942 APOE BURDEN_0.050 2.83457e-72 0.05 0.05 19:45412079:C:T 0.0413056 -0.554561 2.83457e-72
- According to the example above, PCSK9 had a p-value of 7.54587e-11 from the gene-based burden test, where three variants from this gene were included. Another hit from this meta analysis is APOE, where only one variant was included in the burden test.
PLOTS
RAREMETAL generates QQ plots and manhattan plots from single variant and gene-level meta-analysis by default. Example QQ plots and manhattan plots are:
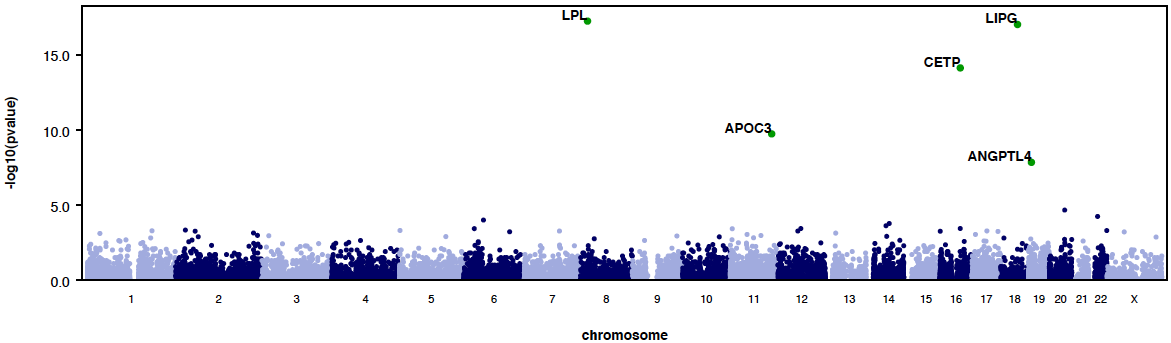
|
LOG
- A log file is automatically generated by RAREMETAL to save the parameters in effect. An example is in the following:
The following parameters are in effect: List of Studies: ============================ --studyName [studyName.SardiNia] Grouping Methods: ============================ --groupFile [genes.file] --annotatedVcf [] --annotation [] --writeVcf [OFF] QC Options: ============================ --hwe [0] --callRate [0] Association Methods: ============================ --burden [true] --MB [false] --SKAT [false] --VT [false] --condition [condition.file] Other Options: ============================ --tabix [OFF] --correctGC [ON] --prefix [test] --maf [0.05] --longOutput [false] --tabulateHits [false] --hitsCutoff [1e-06] --dosage [false] --altMAF [false]
Example Command lines
- Here is an example command line to do single variant meta analysis only:
./raremetal --summaryFiles your.list.of.summary.files --prefix yourPrefix
- When you want to do all burden tests using a group file to specify which variants to group:
./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --burden --MB --SKAT --VT --maf 0.01 --prefix yourPrefix (NOTE: this will generate single variant meta analysis result and the short format output for burden test results.)
- Here is how to do all SKAT meta analysis using a group file and request a long format output together with tabulated hits:
./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --prefix yourPrefix
- Here is an example of adding QC filters to variants when doing meta analysis.
./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --groupFile your.groupfile --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --hwe 1e-06 --callRate 0.98 --prefix yourPrefix
- Here is how to do the same thing but reading grouping information from an annotated VCF file:
./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --annotatedVcf your.annotated.vcf --annotation nonsyn/stop/splicing --SKAT --longOutput --tabulateHits --hitsCutoff 1.0e-07 --hwe 1e-06 --callRate 0.98 --prefix yourPrefix
- If you want to write a VCF file of pooled variants from all studies, annotate them using your favorite annotation program, and then come back to RAREMETAL with the annotate VCF file to do burden tests:
First, use the following command to write the VCF file: ./raremetal --summaryFiles your.list.of.summary.files --writeVcf --prefix yourPrefix Second, annotate the VCF file using your favorite annotation program. (Annotated VCF file has to follow the format described here: annotated VCF format) Third, use the following command to do meta analysis: ./raremetal --summaryFiles your.list.of.summary.files --covFiles your.list.of.cov.files --annotatedVcf your.annotated.vcf --annotation nonsyn/splicing/stop --burden --MB --SKAT --VT --maf 0.01 --prefix yourPrefix
Other Useful Info
- Summary specs can be found Summary Files Specification for RAREMETAL
TUTORIAL
- For a comprehensive tutorial of RAREMETALWORKER and RAREMETAL using example data sets, please go to the following:
RAREMETAL Tutorial
- For a brief tutorial of rvtests, please go to:
rvtests
CONTACT
Please email Andy Boughton (abought at umich dot edu) for questions.
Also check Known issues and incoming update in next version to see if your problem has been reported before