Difference between revisions of "Regions of high linkage disequilibrium (LD)"
From Genome Analysis Wiki
Jump to navigationJump to search| Line 2: | Line 2: | ||
[[Image:High-ld.png]] | [[Image:High-ld.png]] | ||
| + | |||
| + | Here is a lost of positions for GRCH Build 37 | ||
| + | <tab border="1" head="top"> | ||
| + | Chr Start Stop ID | ||
| + | 1 48000000 52000000 | ||
| + | 2 86000000 100500000 | ||
| + | 2 134500000 138000000 | ||
| + | 2 183000000 190000000 | ||
| + | 3 47500000 50000000 | ||
| + | 3 83500000 87000000 | ||
| + | 3 89000000 97500000 | ||
| + | 5 44500000 50500000 | ||
| + | 5 98000000 100500000 | ||
| + | 5 129000000 132000000 | ||
| + | 5 135500000 138500000 | ||
| + | 6 25000000 35000000 | ||
| + | 6 57000000 64000000 | ||
| + | 6 140000000 142500000 | ||
| + | 7 55000000 66000000 | ||
| + | 8 7000000 13000000 | ||
| + | 8 43000000 50000000 | ||
| + | 8 112000000 115000000 | ||
| + | 10 37000000 43000000 | ||
| + | 11 46000000 57000000 | ||
| + | 11 87500000 90500000 | ||
| + | 12 33000000 40000000 | ||
| + | 12 109500000 112000000 | ||
| + | 20 32000000 34500000 | ||
| + | </tab> | ||
| + | |||
These positions are for GRCH build 36. | These positions are for GRCH build 36. | ||
| Line 30: | Line 60: | ||
12 109521663 112021663 hild23 | 12 109521663 112021663 hild23 | ||
20 32000000 34500000 hild24 | 20 32000000 34500000 hild24 | ||
| − | + | X 14150264 16650264 hild25 | |
| − | + | X 25650264 28650264 hild26 | |
| − | + | X 33150264 35650264 hild27 | |
| − | + | X 55133704 60500000 hild28 | |
| − | + | X 65133704 67633704 hild29 | |
| − | + | X 71633704 77580511 hild30 | |
| − | + | X 80080511 86080511 hild31 | |
| − | + | X 100580511 103080511 hild32 | |
| − | + | X 125602146 128102146 hild33 | |
| − | + | X 129102146 131602146 hild34 | |
</tab> | </tab> | ||
Revision as of 13:02, 27 July 2017
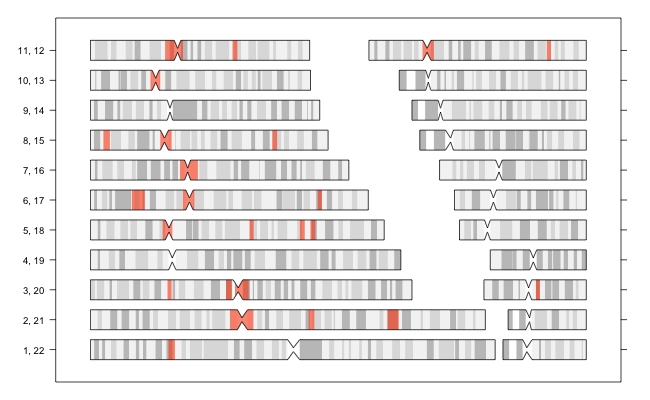
There are regions of long-range, high linkage diequilibrium in the human genome [1][2]. These regions should be excluded when performing certain analyses such as principal component analysis on genotype data.
Here is a lost of positions for GRCH Build 37
| Chr | Start | Stop | ID |
|---|---|---|---|
| 1 | 48000000 | 52000000 | |
| 2 | 86000000 | 100500000 | |
| 2 | 134500000 | 138000000 | |
| 2 | 183000000 | 190000000 | |
| 3 | 47500000 | 50000000 | |
| 3 | 83500000 | 87000000 | |
| 3 | 89000000 | 97500000 | |
| 5 | 44500000 | 50500000 | |
| 5 | 98000000 | 100500000 | |
| 5 | 129000000 | 132000000 | |
| 5 | 135500000 | 138500000 | |
| 6 | 25000000 | 35000000 | |
| 6 | 57000000 | 64000000 | |
| 6 | 140000000 | 142500000 | |
| 7 | 55000000 | 66000000 | |
| 8 | 7000000 | 13000000 | |
| 8 | 43000000 | 50000000 | |
| 8 | 112000000 | 115000000 | |
| 10 | 37000000 | 43000000 | |
| 11 | 46000000 | 57000000 | |
| 11 | 87500000 | 90500000 | |
| 12 | 33000000 | 40000000 | |
| 12 | 109500000 | 112000000 | |
| 20 | 32000000 | 34500000 |
These positions are for GRCH build 36.
| Chr | Start | Stop | ID |
|---|---|---|---|
| 1 | 48060567 | 52060567 | hild1 |
| 2 | 85941853 | 100407914 | hild2 |
| 2 | 134382738 | 137882738 | hild3 |
| 2 | 182882739 | 189882739 | hild4 |
| 3 | 47500000 | 50000000 | hild5 |
| 3 | 83500000 | 87000000 | hild6 |
| 3 | 89000000 | 97500000 | hild7 |
| 5 | 44500000 | 50500000 | hild8 |
| 5 | 98000000 | 100500000 | hild9 |
| 5 | 129000000 | 132000000 | hild10 |
| 5 | 135500000 | 138500000 | hild11 |
| 6 | 25500000 | 33500000 | hild12 |
| 6 | 57000000 | 64000000 | hild13 |
| 6 | 140000000 | 142500000 | hild14 |
| 7 | 55193285 | 66193285 | hild15 |
| 8 | 8000000 | 12000000 | hild16 |
| 8 | 43000000 | 50000000 | hild17 |
| 8 | 112000000 | 115000000 | hild18 |
| 10 | 37000000 | 43000000 | hild19 |
| 11 | 46000000 | 57000000 | hild20 |
| 11 | 87500000 | 90500000 | hild21 |
| 12 | 33000000 | 40000000 | hild22 |
| 12 | 109521663 | 112021663 | hild23 |
| 20 | 32000000 | 34500000 | hild24 |
| X | 14150264 | 16650264 | hild25 |
| X | 25650264 | 28650264 | hild26 |
| X | 33150264 | 35650264 | hild27 |
| X | 55133704 | 60500000 | hild28 |
| X | 65133704 | 67633704 | hild29 |
| X | 71633704 | 77580511 | hild30 |
| X | 80080511 | 86080511 | hild31 |
| X | 100580511 | 103080511 | hild32 |
| X | 125602146 | 128102146 | hild33 |
| X | 129102146 | 131602146 | hild34 |
Excluding Regions With Plink
You can remove these regions from a PED file using the following PLINK commands. Assuming you have the data stored in a file named "high-ld.txt"
plink --file mydata --make-set high-ld.txt --write-set --out hild plink --file mydata --exclude hild.set --recode --out mydatatrimmed
References
- ↑ Price et al. (2008) Long-Range LD Can Confound Genome Scans in Admixed Populations. Am. J. Hum. Genet. 86, 127-147
- ↑ Weale M. (2010) Quality Control for Genome-Wide Association Studies from Michael R. Barnes and Gerome Breen (eds.), Genetic Variation: Methods and Protocols, Methods in Molecular Biology, vol. 628, DOI 10.1007/978-1-60327-367-1_19, © Springer Science+Business Media, LLC 2010