Difference between revisions of "SeqShop: Aligning Your Own Genome, December 2014"
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | == Viewing Genetic Info == | ||
| + | Slides on [[Media:Sequence Analysis Workshop-Risks viewing genetic information Dec2014.pdf|risks of viewing genetic information]] | ||
| + | |||
== Goals of This Session == | == Goals of This Session == | ||
Learn how to go from your FASTQ files to generate Aligned BAMs. | Learn how to go from your FASTQ files to generate Aligned BAMs. | ||
| Line 5: | Line 8: | ||
* You will get to take home both the original FASTQs and the aligned BAMs on a USB drive | * You will get to take home both the original FASTQs and the aligned BAMs on a USB drive | ||
** You will get it by the end of the week if not before - it takes a while to copy 74G-111G | ** You will get it by the end of the week if not before - it takes a while to copy 74G-111G | ||
| − | ** 4 samples will get | + | ** 4 samples will get "modified" BAMs so both FASTQ & BAM would fit on the drive |
| − | *** In future all generated BAMs will be automatically binned | + | *** "binned" qualities, duplicates & unmapped reads removed |
| + | **** In future all generated BAMs will be automatically binned | ||
** Those that didn't get sequenced will get a drive with NA12878 public sample on it | ** Those that didn't get sequenced will get a drive with NA12878 public sample on it | ||
| + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | ||
| + | ''Login instructions for seqshop-server'' | ||
| + | <div class="mw-collapsible-content"> | ||
{{SeqShopLogin}} | {{SeqShopLogin}} | ||
| + | </div> | ||
| + | </div> | ||
| − | == | + | == First Things First == |
| − | + | When you see Sample*/Sample#, replace it with your sample name/number | |
| − | * | + | * If you are using generic data, use NA12878 |
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | == Locating your FASTQs == |
| − | + | Your FASTQ files are under <code>~/Sample*/fastqs</code> directory. | |
| − | Your FASTQ files are under | ||
Look at your directory: | Look at your directory: | ||
| − | ls ~/ | + | ls ~/Sample*/fastqs |
| + | |||
* <code>ls</code> does a directory listing | * <code>ls</code> does a directory listing | ||
* <code>~/</code> means to start from your home/base directory | * <code>~/</code> means to start from your home/base directory | ||
** Using ~/ means the command will work even if you have changed directories | ** Using ~/ means the command will work even if you have changed directories | ||
| − | You | + | You will see 2 files: |
| − | * | + | * Sample#_R1.fastq.gz - 1st in pair |
| + | * Sample#_R2.fastq.gz - 2nd in pair | ||
| − | What | + | == What did I do to run the alignment? == |
| − | + | === Created FASTQ_LIST file === | |
| − | * | + | I created a FASTQ_LIST file with the sample #s and fastqs. |
| − | ** | + | * What columns do we need in our file that tells GotCloud about our FASTQ? |
| + | ** SAMPLE | ||
| + | ** FASTQ1 | ||
| + | ** FASTQ2 | ||
| + | |||
| + | SAMPLE FASTQ1 FASTQ2 | ||
| + | Sample# /path/to/Sample#/fastqs/Sample#_R1.fastq.gz /path/to/Sample#/fastqs/Sample#_R2.fastq.gz | ||
| − | + | You can see what your fastq.list looks like | |
| + | * I took out the full path since I ran alignment in a different path: | ||
| + | cat ~/Sample*/fastq.list | ||
| − | + | === Created a GotCloud Configuration File === | |
| + | After creating the FASTQ_LIST file, I created the GotCloud configuration. | ||
| + | * What changes from the default settings did I make? | ||
| + | *# Use BWA instead of BWA_MEM | ||
| + | *# Use multiple BWA_THREADS | ||
| + | *# Set FASTQ_LIST | ||
| − | + | Look at the gotcloud.conf I setup for you | |
| − | + | * Not "exactly" what I used. In the original gotcloud.conf, I had | |
| − | + | ** cluster settings (now blank since you won't be using a cluster for further processing) | |
| + | ** various number of BWA_THREADS for each sample | ||
| + | ** full paths to: | ||
| + | *** FASTQ_LIST (and I sometimes had multiple samples in 1 list - but each gets aligned independently) | ||
| + | *** OUT_DIR | ||
| + | cat ~/Sample*/gotcloud.conf | ||
| − | + | You'll notice that this file is very similar to the one we have been using. | |
| + | * Just a few modifications to run a new test on the whole genome | ||
| + | The settings needed for Single Sample SNP calling that we need for tomorrow are already in the gotcloud.conf (requires extra settings as the default snpcall works best for multiple samples). | ||
| − | + | === Ran the Alignment === | |
| + | It took many threads & a couple of days to get all of the alignments complete - which is why I ran them last week. | ||
| + | * Used screen to run overnight. | ||
| − | + | I ran something like: | |
| + | gotcloud align --conf gotcloud.conf --numjobs 4 | ||
| + | * I set numjobs to the number of samples I was processing on that machine | ||
| − | == | + | == Alignment Pipeline Output == |
| − | + | The output is in ~/Sample#/output | |
| + | cd ~/Sample*/output | ||
| + | ls | ||
| − | + | What is there? | |
| − | + | * '''bam.list''' - list of samples/BAMs (just one) | |
| + | * '''bams''' - directory with BAM files | ||
| + | * Makefiles - makefiles generated when I ran GotCloud | ||
| + | * QCFiles - quality control metrics | ||
| + | ** We will look at this later | ||
| − | + | Look at the BAM list (will be used for snpcall that we will start tomorrow) | |
| + | cat bam.list | ||
| + | * sample(tab)bam | ||
| − | + | Look in the bams directory: | |
| − | + | ls bams | |
| − | === | + | === Quality Control Output === |
| − | + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | |
| − | + | ''We may hold off on reviewing this until Friday.'' | |
| − | + | <div class="mw-collapsible-content"> | |
| − | + | Check QC directory | |
| − | + | ls QCFiles/ | |
| − | |||
| − | |||
| − | |||
| − | + | Check for Sample Contamination: | |
| + | less -S QCFiles/Sample*.genoCheck.selfSM | ||
| + | Look for FREEMIX column. OR notice that it is column 7: | ||
| + | cut -f7 QCFiles/Sample*.genoCheck.selfSM | ||
| − | + | Look at QPLOT stats: | |
| − | + | less QCFiles/Sample*.qplot.stats | |
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | + | * What is your Mapping Rate%? | |
| − | + | * What is your MeanDepth? | |
| − | * | + | * What is your GenomeCover(%)? |
| − | + | Let's generate the plots: | |
| + | * R script will create PDF | ||
| + | ** automatically set PDF path to full path where the R script is | ||
| + | *** That wouldn't work since I didn't align in your directory & instead moved the files in there afterwards | ||
| + | *** I hand modified it to relative directory from your home directory, so you need to move to your home directory to create the PDF | ||
| + | cd | ||
| + | Rscript Sample*/output/QCFiles/Sample*.qplot.R | ||
| + | evince Sample*/output/QCFiles/Sample*.qplot.pdf& | ||
| + | </div> | ||
| + | </div> | ||
| − | ===== | + | == Recalibration Comparison == |
| − | + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | |
| − | + | ''We may hold off on reviewing this until Friday.'' | |
| − | + | <div class="mw-collapsible-content"> | |
| − | + | I also ran picard/GATK on NA12878. | |
| − | + | {| class="wikitable" cellpadding=5 | |
| − | == | + | ! Tool !! Time |
| − | + | |- | |
| − | + | | Picard MarkDuplicates || 5hrs 41min | |
| − | + | |- | |
| − | + | | GATK BaseRecalibrator || 18hrs 57min | |
| − | + | |- | |
| − | + | | GATK PrintReads || 18hrs 33min | |
| − | + | |- | |
| − | + | ! Picard/GATK Total || 43hrs 11min | |
| − | + | |- | |
| − | + | ! Our Dedup & Recalibration || 15hrs 3min | |
| − | + | |- | |
| − | + | | Just Dedup || 5hr 19min | |
| − | + | |- | |
| − | + | | Just Recalibration || 13hrs 5 min | |
| − | * | + | |} |
| − | + | We run Dedup & Recalibration at the same time for 2 total passes through the BAM file. | |
| − | + | * Alternatively you can run them separately | |
| − | + | Our samples ranged from 8-19 hrs (only 2 at 19-19) | |
| − | + | QPLOT comparison: | |
| + | * qplot.stats differences: | ||
| + | {| class="wikitable" cellpadding=5 | ||
| + | ! Stats\BAM !! NA12878.recal.bam !! NA12878.markDup_GATK.bam | ||
| + | |- | ||
| + | | Q20Bases(e9) || 54.29 || 54.20 | ||
| + | |- | ||
| + | | Q20BasesPct(%) || 94.20 || 94.05 | ||
| + | |- | ||
| + | | EPS_MSE || 3.80 || 1.37 | ||
| + | |} | ||
| − | + | Plots: [[Media:QplotComp.pdf|QplotComp.pdf]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== FEEDBACK! == | == FEEDBACK! == | ||
| − | + | Please provide feedback on the lectures/tutorials from today: https://docs.google.com/forms/d/1HY9g_GzTZzwddA9bmoyG1IJBgKukk3ouVR5uBboydec/viewform | |
| − | https://docs.google.com/forms/d/ | ||
Latest revision as of 10:23, 8 December 2014
Viewing Genetic Info
Slides on risks of viewing genetic information
Goals of This Session
Learn how to go from your FASTQ files to generate Aligned BAMs.
- Your samples have already been aligned, so we will review the steps that were done
- Workshop computers don't have enough compute to align everyone's samples during the workshop
- You will get to take home both the original FASTQs and the aligned BAMs on a USB drive
- You will get it by the end of the week if not before - it takes a while to copy 74G-111G
- 4 samples will get "modified" BAMs so both FASTQ & BAM would fit on the drive
- "binned" qualities, duplicates & unmapped reads removed
- In future all generated BAMs will be automatically binned
- "binned" qualities, duplicates & unmapped reads removed
- Those that didn't get sequenced will get a drive with NA12878 public sample on it
Login instructions for seqshop-server
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
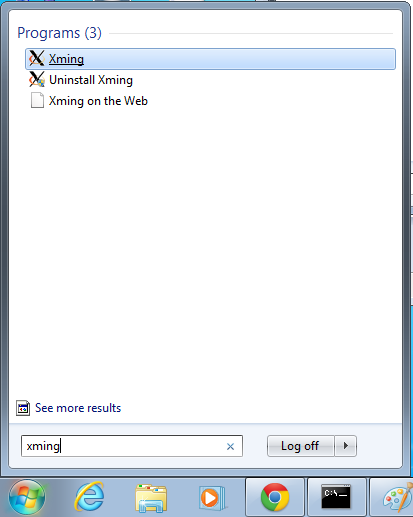
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
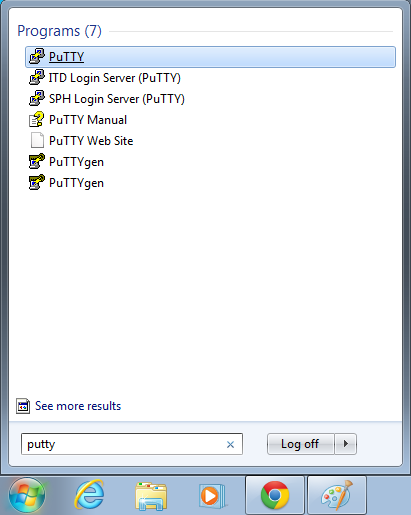
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
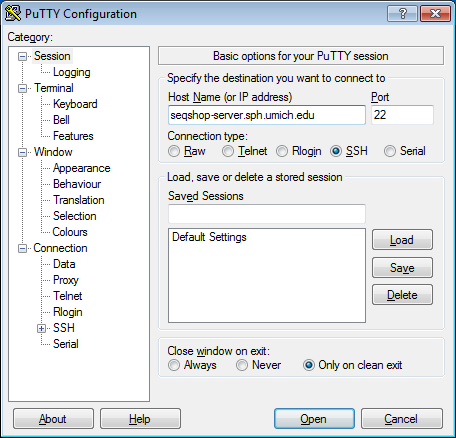
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
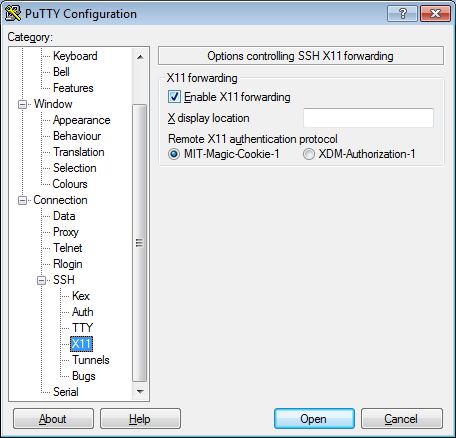
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
First Things First
When you see Sample*/Sample#, replace it with your sample name/number
- If you are using generic data, use NA12878
Locating your FASTQs
Your FASTQ files are under ~/Sample*/fastqs directory.
Look at your directory:
ls ~/Sample*/fastqs
lsdoes a directory listing~/means to start from your home/base directory- Using ~/ means the command will work even if you have changed directories
You will see 2 files:
- Sample#_R1.fastq.gz - 1st in pair
- Sample#_R2.fastq.gz - 2nd in pair
What did I do to run the alignment?
Created FASTQ_LIST file
I created a FASTQ_LIST file with the sample #s and fastqs.
- What columns do we need in our file that tells GotCloud about our FASTQ?
- SAMPLE
- FASTQ1
- FASTQ2
SAMPLE FASTQ1 FASTQ2 Sample# /path/to/Sample#/fastqs/Sample#_R1.fastq.gz /path/to/Sample#/fastqs/Sample#_R2.fastq.gz
You can see what your fastq.list looks like
- I took out the full path since I ran alignment in a different path:
cat ~/Sample*/fastq.list
Created a GotCloud Configuration File
After creating the FASTQ_LIST file, I created the GotCloud configuration.
- What changes from the default settings did I make?
- Use BWA instead of BWA_MEM
- Use multiple BWA_THREADS
- Set FASTQ_LIST
Look at the gotcloud.conf I setup for you
- Not "exactly" what I used. In the original gotcloud.conf, I had
- cluster settings (now blank since you won't be using a cluster for further processing)
- various number of BWA_THREADS for each sample
- full paths to:
- FASTQ_LIST (and I sometimes had multiple samples in 1 list - but each gets aligned independently)
- OUT_DIR
cat ~/Sample*/gotcloud.conf
You'll notice that this file is very similar to the one we have been using.
- Just a few modifications to run a new test on the whole genome
The settings needed for Single Sample SNP calling that we need for tomorrow are already in the gotcloud.conf (requires extra settings as the default snpcall works best for multiple samples).
Ran the Alignment
It took many threads & a couple of days to get all of the alignments complete - which is why I ran them last week.
- Used screen to run overnight.
I ran something like:
gotcloud align --conf gotcloud.conf --numjobs 4
- I set numjobs to the number of samples I was processing on that machine
Alignment Pipeline Output
The output is in ~/Sample#/output
cd ~/Sample*/output ls
What is there?
- bam.list - list of samples/BAMs (just one)
- bams - directory with BAM files
- Makefiles - makefiles generated when I ran GotCloud
- QCFiles - quality control metrics
- We will look at this later
Look at the BAM list (will be used for snpcall that we will start tomorrow)
cat bam.list
- sample(tab)bam
Look in the bams directory:
ls bams
Quality Control Output
We may hold off on reviewing this until Friday.
Check QC directory
ls QCFiles/
Check for Sample Contamination:
less -S QCFiles/Sample*.genoCheck.selfSM
Look for FREEMIX column. OR notice that it is column 7:
cut -f7 QCFiles/Sample*.genoCheck.selfSM
Look at QPLOT stats:
less QCFiles/Sample*.qplot.stats
- What is your Mapping Rate%?
- What is your MeanDepth?
- What is your GenomeCover(%)?
Let's generate the plots:
- R script will create PDF
- automatically set PDF path to full path where the R script is
- That wouldn't work since I didn't align in your directory & instead moved the files in there afterwards
- I hand modified it to relative directory from your home directory, so you need to move to your home directory to create the PDF
- automatically set PDF path to full path where the R script is
cd Rscript Sample*/output/QCFiles/Sample*.qplot.R evince Sample*/output/QCFiles/Sample*.qplot.pdf&
Recalibration Comparison
We may hold off on reviewing this until Friday.
I also ran picard/GATK on NA12878.
| Tool | Time |
|---|---|
| Picard MarkDuplicates | 5hrs 41min |
| GATK BaseRecalibrator | 18hrs 57min |
| GATK PrintReads | 18hrs 33min |
| Picard/GATK Total | 43hrs 11min |
| Our Dedup & Recalibration | 15hrs 3min |
| Just Dedup | 5hr 19min |
| Just Recalibration | 13hrs 5 min |
We run Dedup & Recalibration at the same time for 2 total passes through the BAM file.
- Alternatively you can run them separately
Our samples ranged from 8-19 hrs (only 2 at 19-19)
QPLOT comparison:
- qplot.stats differences:
| Stats\BAM | NA12878.recal.bam | NA12878.markDup_GATK.bam |
|---|---|---|
| Q20Bases(e9) | 54.29 | 54.20 |
| Q20BasesPct(%) | 94.20 | 94.05 |
| EPS_MSE | 3.80 | 1.37 |
Plots: QplotComp.pdf
FEEDBACK!
Please provide feedback on the lectures/tutorials from today: https://docs.google.com/forms/d/1HY9g_GzTZzwddA9bmoyG1IJBgKukk3ouVR5uBboydec/viewform