SeqShop: Aligning Your Own Genome, December 2014
Goals of This Session
Learn how to go from your FASTQ files to generate Aligned BAMs.
- Your samples have already been aligned, so we will review the steps that were done
- Workshop computers don't have enough compute to align everyone's samples during the workshop
- You will get to take home both the original FASTQs and the aligned BAMs on a USB drive
- You will get it by the end of the week if not before - it takes a while to copy 74G-111G
- 4 samples will get BAMs with "binned" qualities so both FASTQ & BAM would fit on the drive
- In future all generated BAMs will be automatically binned.
- Those that didn't get sequenced will get a drive with NA12878 public sample on it
Login instructions for seqshop-server
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
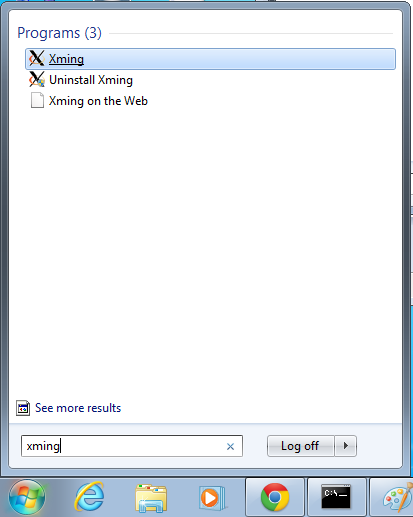
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
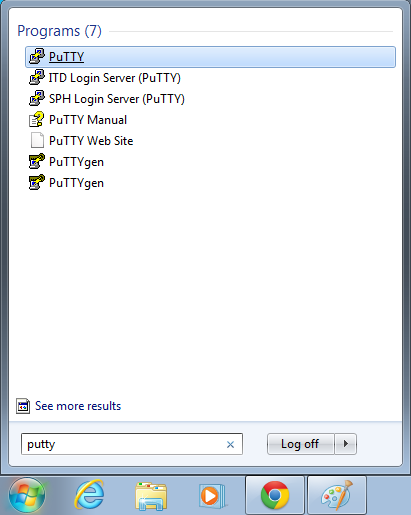
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
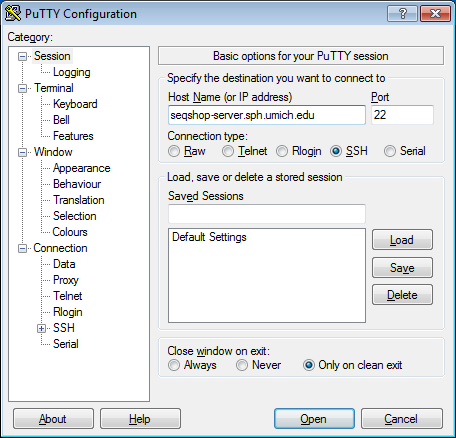
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
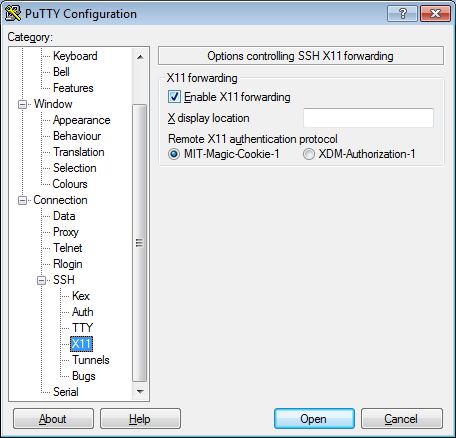
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
I got sequence, how to I get my data ready to run?
Finding your FASTQs
Your FASTQ files are under your personal directory.
Look at your directory:
ls ~/personal
lsdoes a directory listing~/means to start from your home/base directory- Using ~/ means the command will work even if you have changed directories
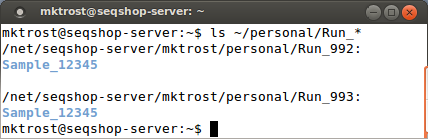
You should see 2 directories: Run_992 & Run_993
- Our DNA was run through as 2 sequencing runs.
What is under those directories? Let's check:
ls ~/personal/Run_*
*is a Linux wild-card match- It will do a directory listing on both Run directories at the same time:
You will see your sample name instead of 12345.
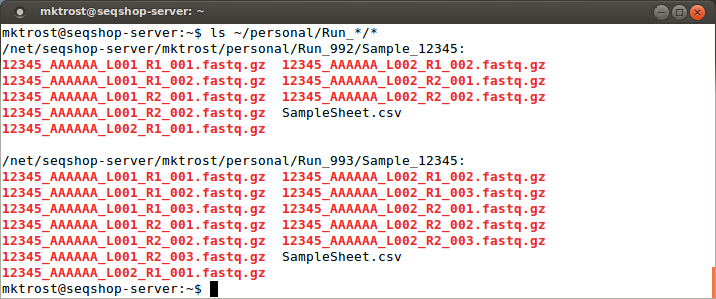
Since we found another directory, let's check below that for our fastqs:
ls ~/personal/Run_*/*
You should see your fastq files:
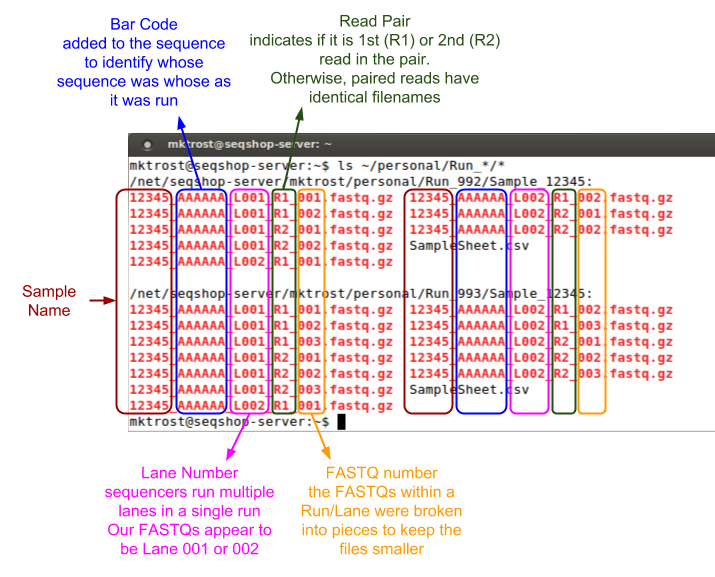
Those filenames are cryptic, what do they mean?
Checking your index file listing your FASTQs
Are you analyzing your own genome? Do you think you setup your file correctly?
Try running this script to see if you have any errors:
perl /home/mktrost/seqshop/inputs/checkIndex.pl ~/personal/align.2x.index
On success it prints: Congratulations, your fastq index looks valid
NOTE: This script is tailored to the filenames provided by our sequencing core as described above.
- It could be tailored to other methods, but is designed for the paths of our data.
Generating the index file listing your FASTQs
What columns do we need in our file that tells GotCloud about our FASTQ?
- MERGE_NAME
- FASTQ1
- FASTQ2
- RGID
- SAMPLE
- LIBRARY
- PLATFORM
We will store our FASTQ info file in: ~/personal/align.2x.index.
There are a few ways to create this file.
- Write into a text file one fastq pair at a time.
- Copy fastq1s into a spreadsheet, fill it in and copy back to a text file
- Write/Use a script
Using a Regular Text File
Follow the instructions below, but do it one FASTQ1 at a time (you won't be able to paste a full column of FASTQs at a time).
- Remember to put a tab between each field.
Using a Spreadsheet
Since we just have a handful of FASTQs, we can use a spreadsheet to construct our file and then copy the data into a text file.
- Thanks to those who thought to do this yesterday - it was a great idea.
First, open Excel
Header Row
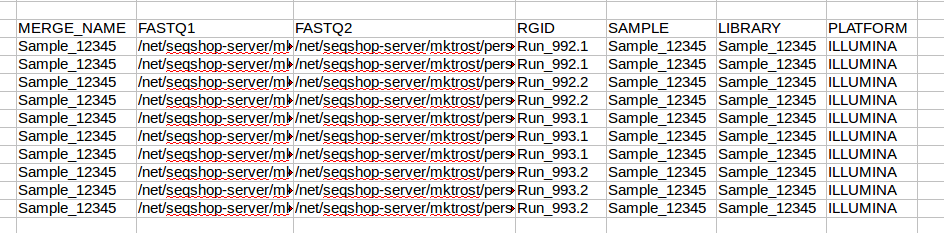
Create the header line by typing each of the column names in a row (you may be able to copy this line):
- make sure you enter these in all CAPS & spelling does matter
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY PLATFORM
MERGE_NAME
MERGE_NAME is just your sample name
- Type your Sample name under the MERGE_NAME column, for example:
Sample_12345
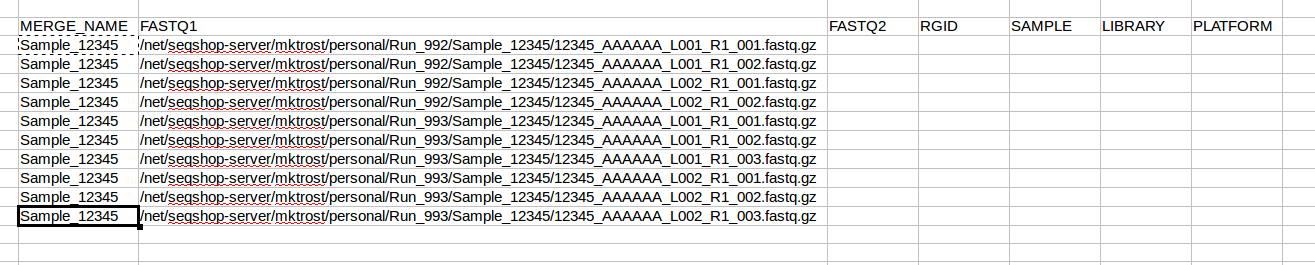
All FASTQs are for the same sample, so you will use Sample_12345 on every line. We will fill those in after we get know how many rows we need.
FASTQ1
FASTQ1 is just the 1st in pair FASTQs (or the single FASTQ in single end)
- Our sequencing core indicated 1st in pair by
R1in the filename.- All our FASTQs are paired end (no single end fastqs)
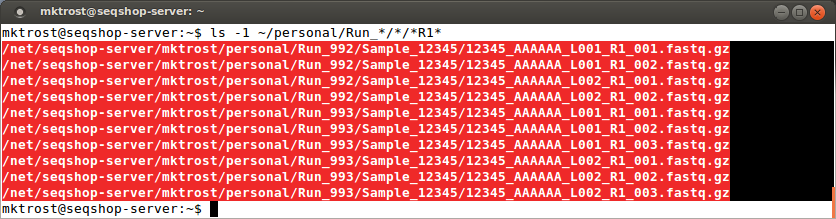
- To get a a list of just the
R1files:
ls -1 ~/personal/Run_*/*/*R1*
- The -1 option tells
lsto list the matching files in a single column- If you have noticed, the number of columns in a directory listing varies based on the width of your window.
- We want to copy these files into one column of a spreadsheet so want them displayed as a single column
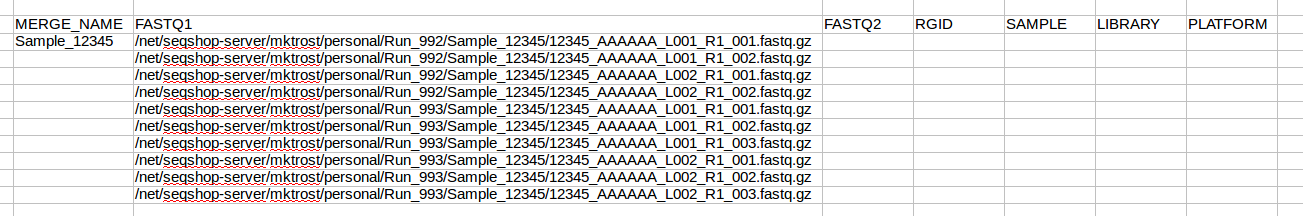
Highlight this list and copy them into the FASTQ1 column of your spreadsheet:
Now that we know how many rows we have, copy your sample name into all rows:
FASTQ2
As mentioned before, FASTQ2 files are the 2nd in pair.
- They have the same filename as FASTQ1, except replace the R1 with R2
You could do an ls and copy & paste as we did for FASTQ1, BUT we'd have to make sure we properly matched up the mates.
EASIER solution:
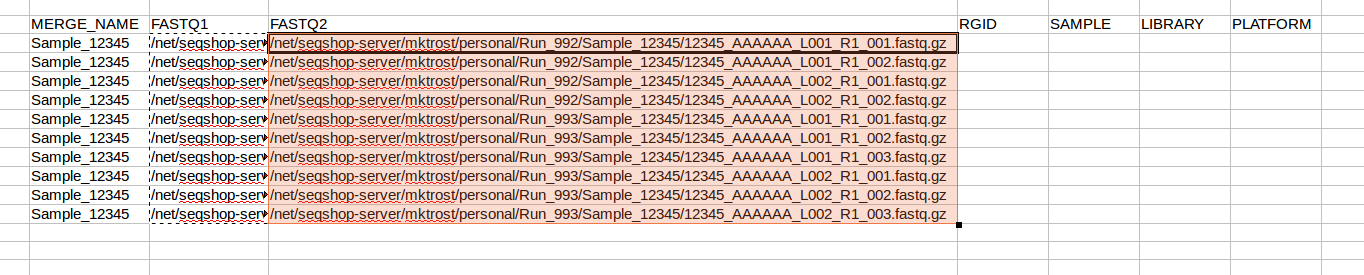
- In your spreadsheet, copy your FASTQ1 filenames into the FASTQ2 column:
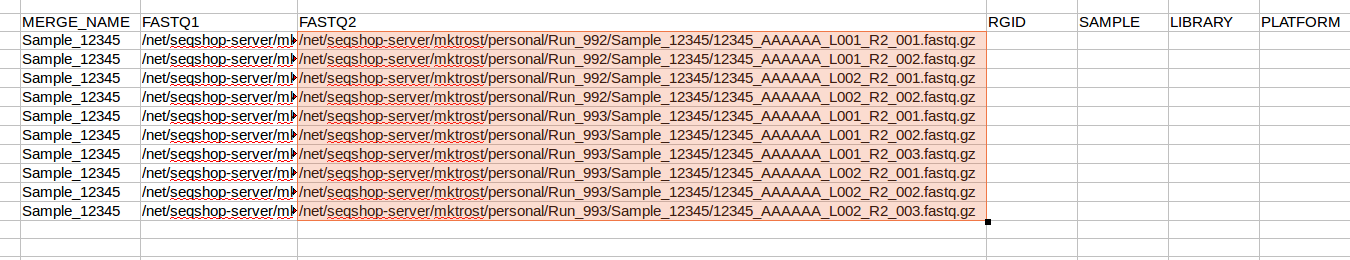
- Now, replace
R1withR2in JUST the FASTQ2 column- Make sure FASTQ1 column keeps the
R1
- Make sure FASTQ1 column keeps the
RGID
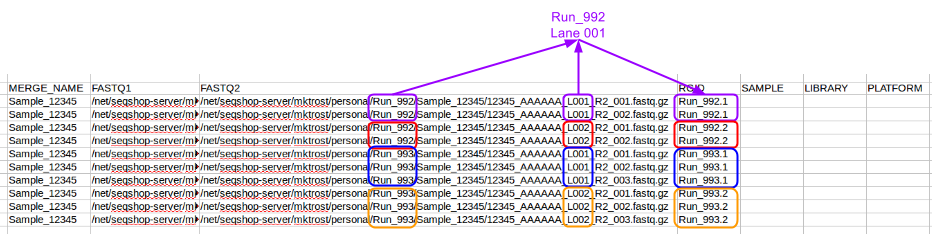
We want to group our FASTQs by Run & Lane.
- Each Run/Lane combination should have a unique Read Group
- Run is in the directory path: Run_992 or Run_993
- Lane is indicated by L001/L002
- Lanes may have multiple FASTQ pairs
- the sequencing core split them into smaller FASTQs, but they are the same Run/Lane, so should have the same read group.
- Lanes may have multiple FASTQ pairs
Populate the RGID column with a name unique to each Read Group/Lane combination:
SAMPLE
Put your sample name in each row of this column (you can copy from MERGE_NAME)
LIBRARY
Put your sample name in each row of this column (you can copy from MERGE_NAME)
- If a sample has multiple library preparations done on it, you would want to give unique names
- That is not our case, so just put in the sample name.
PLATFORM
Your data was sequenced on ILLUMINA, so enter ILLUMINA in each row of the platform column.
Copy to Text File
Open nedit or your favorite linux editor
nedit ~/personal/align.2x.index&
Click New File in the pop-up stating that it couldn't find that file.
Copy (Ctrl-c) your table from EXCEL (including the header row, but with no extra rows/columns.
Paste (Ctrl-v) into your nedit window.
Navigate through the file - the columns should be delimited with tabs.
Save (Ctrl-s) & close nedit.
You now have a tab delimited align.2x.index file (a little simpler than yesterday).
Using a Script
When generating an index of your FASTQs, it can be easiest to have a script.
- Especially if you have many samples/runs, it would be very tedious to do by hand
If you are good at scripting, this may be even easier than doing it by hand
- If you aren't good at scripting, and you have too much data to do by hand
- Make friends with someone who is :-)
- I always find it useful to start from another script (reminds me of commands/tricks)
If you still need to create your file and you don't want to use the spreadsheet method above, you can run a script that I made:
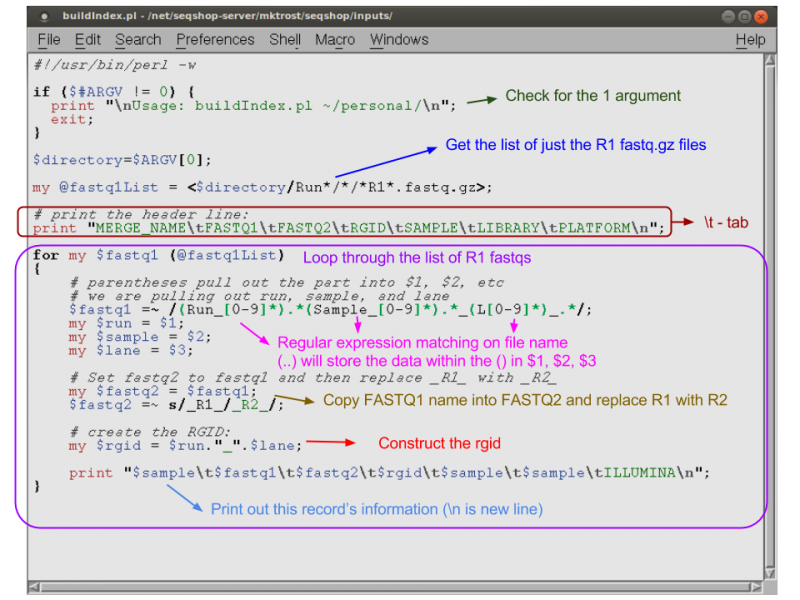
perl /home/mktrost/seqshop/inputs/buildIndex.pl ~/personal > ~/personal/align.2x.index
>means to direct the output to the file specified after the>
Curious what the script looks like and what it does in case you want to create one in the future?
Checking your index file listing your FASTQs
Are you analyzing your own genome? Do you think you setup your file correctly?
Try running this script to see if you have any errors:
perl /home/mktrost/seqshop/inputs/checkIndex.pl ~/personal/align.2x.index
On success it prints: Congratulations, your fastq index looks valid
NOTE: This script is tailored to the filenames provided by our sequencing core as described above.
- It could be tailored to other methods, but is designed for the paths of our data.
Create your GotCloud Configuration File
Now that you have your FASTQ info file created, we need to setup the configuration.
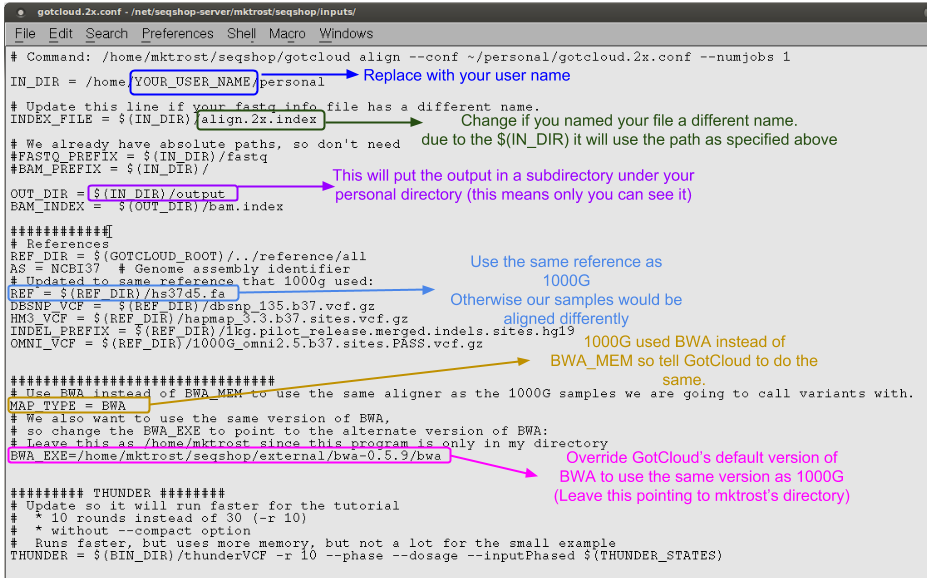
If you updated your gotcloud.2x.conf file yesterday, you still need to do this step
- We updated a few settings to match 1000g
- Updated the version of the genome reference
- Updated to use BWA instead of BWA_MEM
- Updated the version of BWA
You can copy from my directory to your personal directory:
cp /home/mktrost/seqshop/inputs/gotcloud.2x.conf ~/personal/.
You need to update the conf file to find your align.2x.index and ensure your output directory is setup.
nedit ~/personal/gotcloud.2x.conf &
(or use your favorite editor)
- Change the
IN_DIRline, replacingYOUR_USER_NAMEwith your user name. - Everything else is configured already.
You'll notice that this file is very similar to the one we have been using.
- Just a few modifications to run a new test on the whole genome
Run your Alignment
Screen
The alignment pipeline will run overnight, but you'll want to log out.
- How do I leave something running on the server even if I log out?
- One solution is screen!
- How do I use screen?
- Before running your command, you need to start screen:
screen
As it says, press Space or Return.
- It should now look basically the same as your normal command line.
You can now start your alignment:
/home/mktrost/seqshop/gotcloud/gotcloud align --conf ~/personal/gotcloud.2x.conf --numjobs 1
Yes, leave that as /home/mktrost/seqshop/gotcloud/gotcloud - that is where gotcloud is installed. The ~/... points GotCloud to your specific configuration file.
You should now see your alignment running:
- Want to log out and leave your job running?
In the screen window, type:
Ctrl-a d
(Hold down Ctrl and type 'a', let go of both and type 'd')
- This will "detach" from your screen session while your alignment continues to run.
- How do you log back into screen tomorrow?
screen -r
This will resume an already running screen.
- Feel free to test it out and you will see your alignment still running
- Just use Ctrl-a d to detach from screen and leave your job running
- Scrolling problems?
- If you want to scroll and screen doesn't scroll like you normally would?
- Type Ctrl-a Esc and you should be able to scroll up with your mouse wheel
- Or at least that is what I do from my Linux machine - (sorry I'm typing this up/testing these commands from Linux and not windows, so can't test it out)
- Type Ctrl-a Esc and you should be able to scroll up with your mouse wheel
Log Out
If you have not detached from screen:
Ctrl-a d
exit PuTTY
FEEDBACK!
Since I didn't send this out yesterday, today's survey has feedback for Tuesday & Wednesday. https://docs.google.com/forms/d/1qaLHq9w1Ib3FZq0CtlrbK_-breNiqGRV06oRYNmUuME/viewform