Difference between revisions of "SeqShop: Aligning Your Own Genome, June 2014"
| Line 126: | Line 126: | ||
** That is not our case, so just put in the sample name. | ** That is not our case, so just put in the sample name. | ||
| − | ==== PLATFORM | + | ==== PLATFORM ==== |
Your data was sequenced on ILLUMINA, so enter <code>ILLUMINA</code> in each row of the platform column. | Your data was sequenced on ILLUMINA, so enter <code>ILLUMINA</code> in each row of the platform column. | ||
[[File:HdrSheetDone.png]] | [[File:HdrSheetDone.png]] | ||
Revision as of 00:15, 18 June 2014
First Things First
- Helpful reference to many tools:
- http://infoplatter.wordpress.com/2014/04/06/bioinformaticians-pocket-reference/
- links to "cheat-sheets", including, Unix, screen, and vi
- http://infoplatter.wordpress.com/2014/04/06/bioinformaticians-pocket-reference/
- Our wiki with some brief description of how to do some basic commands
- Screen Commands for commands to use screen (one way to leave your commands running even after you log out)
Goals of This Session
Learn how to go from your FASTQ files to generate Aligned BAMs.
- Practice setting up and running GotCloud on your own.
Step 1 : Looking at your FASTQs
Your FASTQ files are under your personal directory.
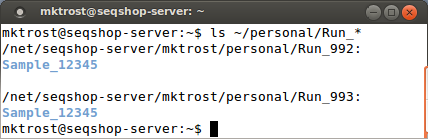
Look at your directory:
ls ~/personal
lsdoes a directory listing~/means to start from your home/base directory- Using ~/ means the command will work even if you have changed directories
You should see 2 directories: Run_992 & Run_993
- Our DNA was run through as 2 sequencing runs.
What is under those directories? Let's check:
ls ~/personal/Run_*
*is a Linux wild-card match- It will do a directory listing on both Run directories at the same time:
You will see your sample name instead of 12345.
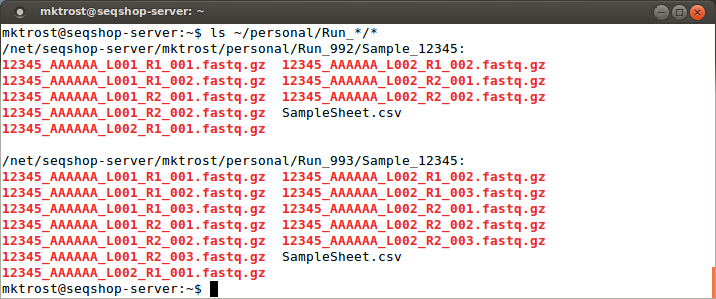
Since we found another directory, let's check below that for our fastqs:
ls ~/personal/Run_*/*
You should see your fastq files:
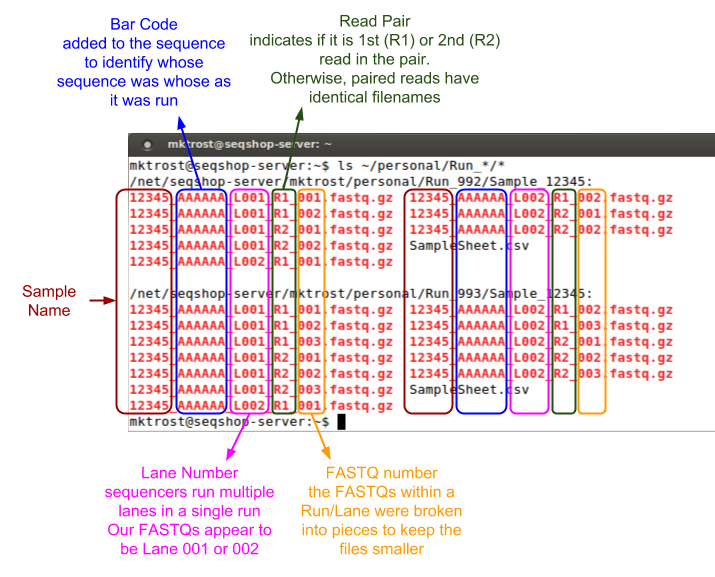
Those filenames are cryptic, what do they mean?
Generating the index file listing your FASTQs
What columns do we need in our file that tells GotCloud about our FASTQ?
- MERGE_NAME
- FASTQ1
- FASTQ2
- RGID
- SAMPLE
- LIBRARY
- PLATFORM
We will store our FASTQ info file in: ~/personal/align.2x.index.
Using a Spreadsheet
Since we just have a handful of FASTQs, we can use a spreadsheet to construct our file and then copy the data into a text file.
- Thanks to those who thought to do this yesterday - it was a great idea.
First, open Excel
Header Row
Create the header line by typing each of the column names in a row:

MERGE_NAME
MERGE_NAME is just your sample name
- Type your Sample name under the MERGE_NAME column, for example:
Sample_12345
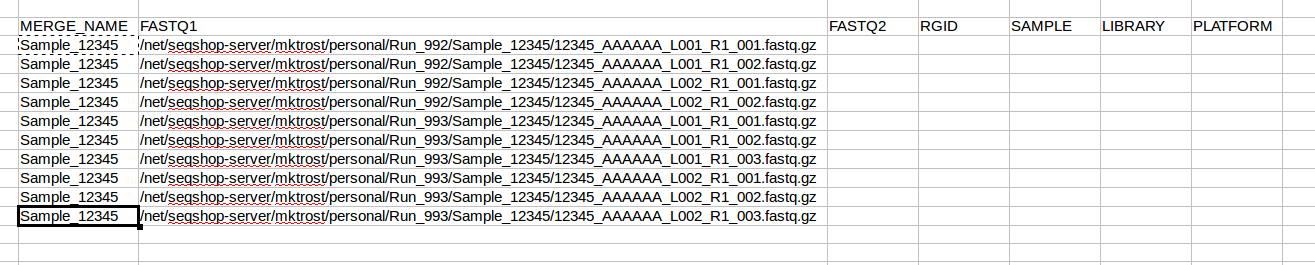
All FASTQs are for the same sample, so you will use Sample_12345 on every line. We will fill those in after we get know how many rows we need.
FASTQ1
FASTQ1 is just the 1st in pair FASTQs (or the single FASTQ in single end)
- Our sequencing core indicated 1st in pair by
R1in the filename.- All our FASTQs are paired end (no single end fastqs)
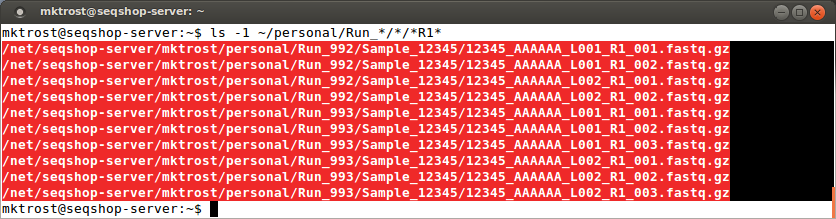
- To get a a list of just the
R1files:
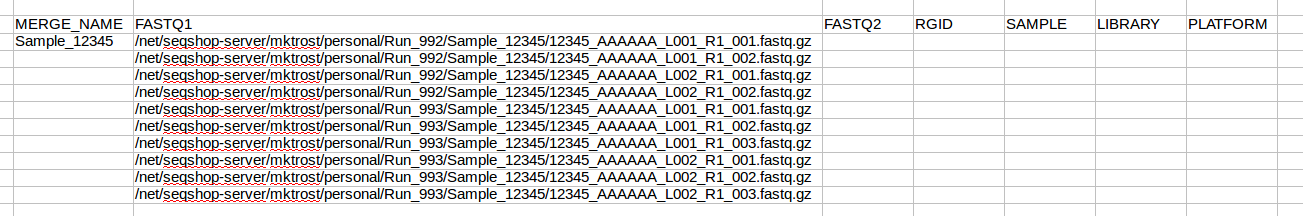
ls -1 ~/personal/Run_*/*/*R1*
- The -1 option tells
lsto list the matching files in a single column- If you have noticed, the number of columns in a directory listing varies based on the width of your window.
- We want to copy these files into one column of a spreadsheet so want them displayed as a single column
Highlight this list and copy them into the FASTQ1 column of your spreadsheet:
Now that we know how many rows we have, copy your sample name into all rows:
FASTQ2
As mentioned before, FASTQ2 files are the 2nd in pair.
- They have the same filename as FASTQ1, except replace the R1 with R2
You could do an ls and copy & paste as we did for FASTQ1, BUT we'd have to make sure we properly matched up the mates.
EASIER solution:
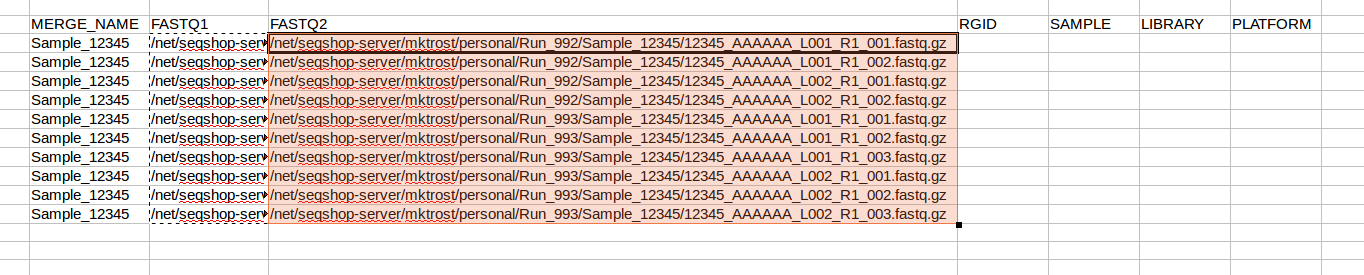
- In your spreadsheet, copy your FASTQ1 filenames into the FASTQ2 column:
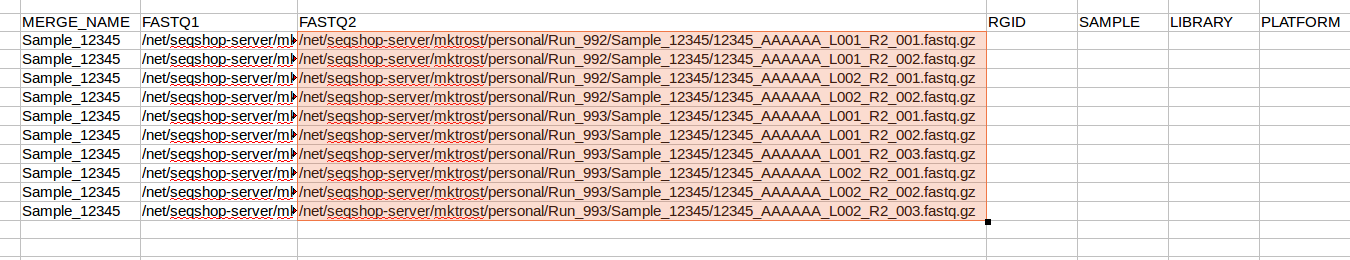
- Now, replace
R1withR2in JUST the FASTQ2 column- Make sure FASTQ1 column keeps the
R1
- Make sure FASTQ1 column keeps the
RGID
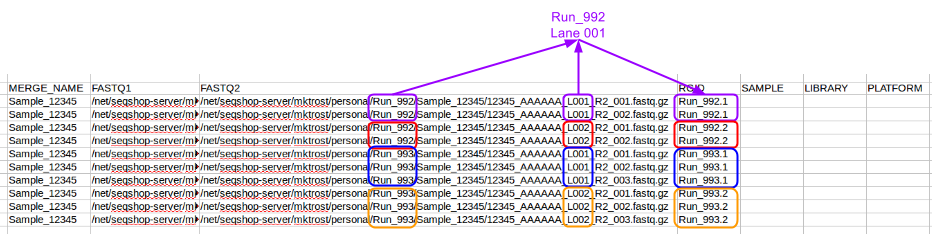
We want to group our FASTQs by Run & Lane.
- Each Run/Lane combination should have a unique Read Group
- Run is in the directory path: Run_992 or Run_993
- Lane is indicated by L001/L002
- Lanes may have multiple FASTQ pairs
- the sequencing core split them into smaller FASTQs, but they are the same Run/Lane, so should have the same read group.
- Lanes may have multiple FASTQ pairs
Populate the RGID column with a name unique to each Read Group/Lane combination:
SAMPLE
Put your sample name in each row of this column (you can copy from MERGE_NAME)
LIBRARY
Put your sample name in each row of this column (you can copy from MERGE_NAME)
- If a sample has multiple library preparations done on it, you would want to give unique names
- That is not our case, so just put in the sample name.
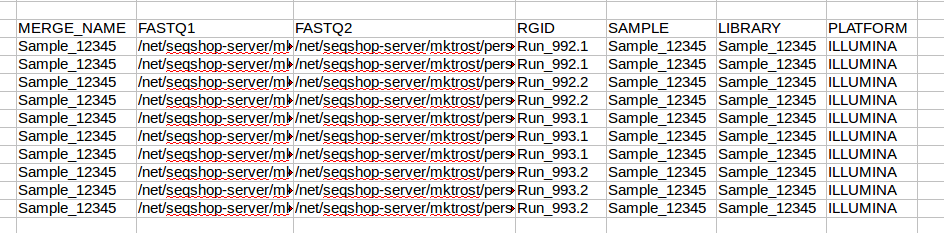
PLATFORM
Your data was sequenced on ILLUMINA, so enter ILLUMINA in each row of the platform column.
Copy to Text File
Open nedit or your favorite linux editor
nedit ~/personal/align.2x.index&
Click New File in the pop-up stating that it couldn't find that file.
Copy (Ctrl-c) your table from EXCEL (including the header row, but with no extra rows/columns.
Paste (Ctrl-v) into your nedit window.
Navigate through the file - the columns should be delimited with tabs.
Save (Ctrl-s) & close nedit.
You now have a tab delimited align.2x.index file (a little simpler than yesterday).