Difference between revisions of "SeqShop: Ancestry On Your Own Genome, December 2014"
| (24 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __TOC__ | ||
| + | |||
| + | == Setup == | ||
| + | <div class="mw-collapsible" style="width:600px"> | ||
| + | ''If you are already logged in, you can skip this section.'' | ||
| + | <div class="mw-collapsible-content"> | ||
{{SeqShopLogin}} | {{SeqShopLogin}} | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | == Setup your terminal == | ||
| − | == | + | Resume screen: |
| − | '' | + | screen -r |
| − | export SAMPLE= | + | |
| + | |||
| + | <div class="mw-collapsible mw-collapsed" style="width:700px"> | ||
| + | '''Only if your snpcalling is still running, let Mary Kate know, expand, and follow these instructions''' | ||
| + | <div class="mw-collapsible-content"> | ||
| + | * Detach from screen | ||
| + | Ctrl-a d | ||
| + | * Start new screen | ||
| + | screen | ||
| + | * Reset environment variables (replace SampleXX with your sample name) | ||
| + | export SAMPLE=SampleXX | ||
source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | ||
| − | + | * When you detach from screen and reattach, you will have multiple screen sessions | |
| + | ** The ancestry one will have today's date, the snpcall one will have an earlier date. | ||
| + | *** Mary Kate can explain how to reattach to a specific session when we get there. | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | '''Instructions for everyone: Setup LASER''' | ||
| − | + | Source the LASER setup: | |
| − | |||
| − | |||
source /net/seqshop-server/home/chaolong/LASER-Tutorial/setup.txt | source /net/seqshop-server/home/chaolong/LASER-Tutorial/setup.txt | ||
| + | (SAMPLE & OUT were previously set in your screen session.) | ||
After setting this, also do | After setting this, also do | ||
| Line 26: | Line 52: | ||
This step takes ~1 hour for a genome sequenced at 17X. | This step takes ~1 hour for a genome sequenced at 17X. | ||
| + | While this is running, we will go look at other results, so exit screen: | ||
| + | Ctrl-a d | ||
| + | |||
| + | While that's running, let's go run the Association Analysis Practical: [[SeqShop: Genetic Association Analysis Practical, December 2014]] | ||
| + | |||
| + | === Checking if Pileup finished === | ||
| + | screen -r | ||
| + | |||
| + | '''Did everyone's finish?''' | ||
| + | |||
| + | |||
| + | If not, let's go to Indel: [[SeqShop:_Calling_Your_Own_Genome,_December_2014#Reviewing_Indel_Results]] | ||
| + | |||
| + | |||
| + | If yes, let's continue below: | ||
=== Step 2: pileup --> seq === | === Step 2: pileup --> seq === | ||
| Line 38: | Line 79: | ||
This step will take about 5-6 minutes. | This step will take about 5-6 minutes. | ||
| − | $LASER/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s $OUT/ancestry/$SAMPLE. | + | $LASER/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP & |
View the results: | View the results: | ||
| Line 56: | Line 97: | ||
evince Results_on_HGDP.pdf & | evince Results_on_HGDP.pdf & | ||
| − | ==Interested in looking just at European | + | ==Interested in looking just at European or East Asian or Central South Asian populations?== |
=== Population information === | === Population information === | ||
8 European populations in the HGDP dataset, including 156 individuals in total. | 8 European populations in the HGDP dataset, including 156 individuals in total. | ||
| − | + | ||
| + | 18 East Asian populations in the HGDP dataset, including 224 individuals after excluding 5 outliers (defined as more than 5 standard deviations from mean in any of top 10 PCs, 1 removal iteration). | ||
| + | |||
8 Central/South Asian populations in the HGDP dataset, including 195 individuals after excluding 5 outliers (defined as more than 5 standard deviations from mean in any of top 10 PCs, 1 removal iteration). Note that 7 out of these 8 population were collected in Pakistan and 1 was collected in China (Uygur). | 8 Central/South Asian populations in the HGDP dataset, including 195 individuals after excluding 5 outliers (defined as more than 5 standard deviations from mean in any of top 10 PCs, 1 removal iteration). Note that 7 out of these 8 population were collected in Pakistan and 1 was collected in China (Uygur). | ||
| + | |||
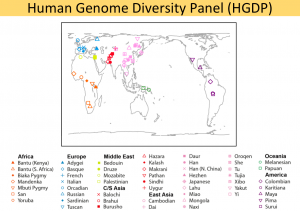
| + | [[File:HGDP Popualtions.png|thumb|center|alt=HGDP populations |300px|HGDP populations]] | ||
=== Step 1: bam --> pileup === | === Step 1: bam --> pileup === | ||
| Line 72: | Line 117: | ||
This step will take a few seconds. | This step will take a few seconds. | ||
| + | For European ancestry: | ||
$LASER/laser -g $HGDP/HGDP.633K.euro.geno -c $HGDP/HGDP.633K.euro.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-Euro & | $LASER/laser -g $HGDP/HGDP.633K.euro.geno -c $HGDP/HGDP.633K.euro.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-Euro & | ||
| − | View the results: | + | For East Asian ancestry: |
| + | $LASER/laser -g $HGDP/HGDP.633K.easia.2.geno -c $HGDP/HGDP.633K.easia.2.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-EAsia & | ||
| + | |||
| + | For Central/South Asian ancestry: | ||
| + | $LASER/laser -g $HGDP/HGDP.633K.csasia.2.geno -c $HGDP/HGDP.633K.csasia.2.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-CSAsia & | ||
| + | |||
| + | |||
| + | View the results for European ancestry: | ||
less -S $OUT/ancestry/${SAMPLE}.HGDP-Euro.SeqPC.coord | less -S $OUT/ancestry/${SAMPLE}.HGDP-Euro.SeqPC.coord | ||
| + | or for East Asian ancestry | ||
| + | less -S $OUT/ancestry/${SAMPLE}.HGDP-EAsia.SeqPC.coord | ||
| + | or for Central/South Asian ancestry | ||
| + | less -S $OUT/ancestry/${SAMPLE}.HGDP-CSAsia.SeqPC.coord | ||
== Visualizing Ancestry == | == Visualizing Ancestry == | ||
| − | |||
| − | |||
Change to that new directory: | Change to that new directory: | ||
| Line 88: | Line 143: | ||
| − | Generate the plot: | + | Generate the plot for European ancestry: |
| − | Rscript plotHGDP.r $HGDP/HGDP.633K.euro.RefPC.coord $OUT/ancestry/${SAMPLE}.Euro. | + | Rscript plotHGDP.r $HGDP/HGDP.633K.euro.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-Euro.SeqPC.coord |
| + | or for East Asian ancestry | ||
| + | Rscript plotHGDP.r $HGDP/HGDP.633K.easia.2.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-EAsia.SeqPC.coord | ||
| + | or for Central/South Asian ancestry | ||
| + | Rscript plotHGDP.r $HGDP/HGDP.633K.csasia.2.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-CSAsia.SeqPC.coord | ||
Take a look: | Take a look: | ||
evince Results_on_HGDP.pdf & | evince Results_on_HGDP.pdf & | ||
| + | |||
| + | == Return to Calling your own Genome == | ||
| + | Go to [[SeqShop:_Calling_Your_Own_Genome,_December_2014#Reviewing_Indel_Results]] and pick up where we left off. | ||
Latest revision as of 12:43, 12 December 2014
Setup
If you are already logged in, you can skip this section.
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
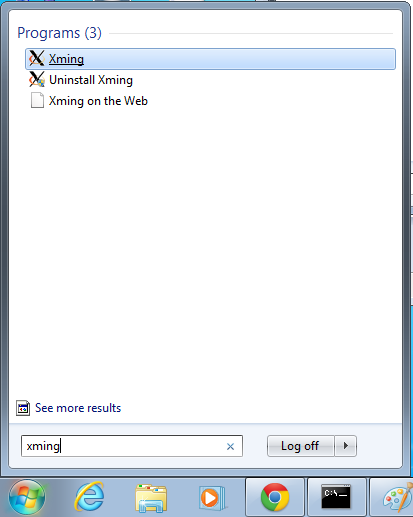
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
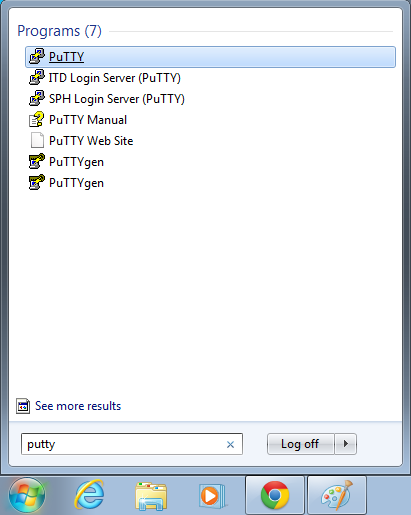
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
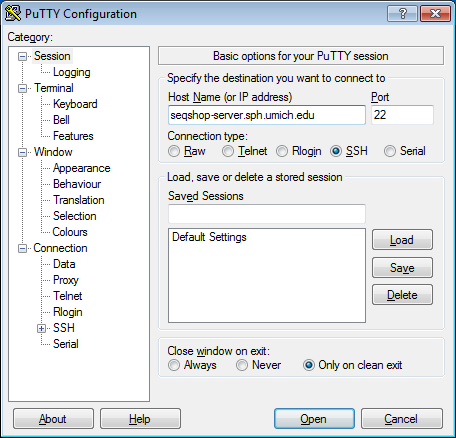
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
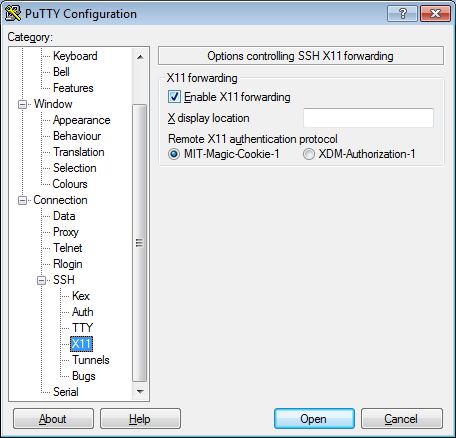
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup your terminal
Resume screen:
screen -r
Only if your snpcalling is still running, let Mary Kate know, expand, and follow these instructions
- Detach from screen
Ctrl-a d
- Start new screen
screen
- Reset environment variables (replace SampleXX with your sample name)
export SAMPLE=SampleXX source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
- When you detach from screen and reattach, you will have multiple screen sessions
- The ancestry one will have today's date, the snpcall one will have an earlier date.
- Mary Kate can explain how to reattach to a specific session when we get there.
- The ancestry one will have today's date, the snpcall one will have an earlier date.
Instructions for everyone: Setup LASER
Source the LASER setup:
source /net/seqshop-server/home/chaolong/LASER-Tutorial/setup.txt
(SAMPLE & OUT were previously set in your screen session.)
After setting this, also do
mkdir -p $OUT/ancestry
Verify that this does not give an error:
ls $OUT/bams/${SAMPLE}.recal.bam
Run
Step 1: bam --> pileup
$GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $OUT/bams/${SAMPLE}.recal.bam > $OUT/ancestry/${SAMPLE}.HGDP.pileup
This step takes ~1 hour for a genome sequenced at 17X.
While this is running, we will go look at other results, so exit screen:
Ctrl-a d
While that's running, let's go run the Association Analysis Practical: SeqShop: Genetic Association Analysis Practical, December 2014
Checking if Pileup finished
screen -r
Did everyone's finish?
If not, let's go to Indel: SeqShop:_Calling_Your_Own_Genome,_December_2014#Reviewing_Indel_Results
If yes, let's continue below:
Step 2: pileup --> seq
python $LASER/pileup2seq/pileup2seq.py \ -m $HGDP/HGDP_938.site \ -o $OUT/ancestry/$SAMPLE.HGDP \ $OUT/ancestry/$SAMPLE.HGDP.pileup
This step takes just a few seconds.
Estimate ancestry
This step will take about 5-6 minutes.
$LASER/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP &
View the results:
less -S $OUT/ancestry/${SAMPLE}.HGDP.SeqPC.coord
Visualizing Ancestry
Copy the R code to plot your ancestry
cp -r $LASER/plot/ $OUT/ancestry/.
Change to that new directory:
cd $OUT/ancestry/plot
Generate the plot:
Rscript plotHGDP.r $HGDP/HGDP_938.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP.SeqPC.coord
Take a look:
evince Results_on_HGDP.pdf &
Interested in looking just at European or East Asian or Central South Asian populations?
Population information
8 European populations in the HGDP dataset, including 156 individuals in total.
18 East Asian populations in the HGDP dataset, including 224 individuals after excluding 5 outliers (defined as more than 5 standard deviations from mean in any of top 10 PCs, 1 removal iteration).
8 Central/South Asian populations in the HGDP dataset, including 195 individuals after excluding 5 outliers (defined as more than 5 standard deviations from mean in any of top 10 PCs, 1 removal iteration). Note that 7 out of these 8 population were collected in Pakistan and 1 was collected in China (Uygur).
Step 1: bam --> pileup
You can skip this, you already did it.
Step 2: pileup --> seq
You can skip this step too, because you already did it.
Estimate ancestry
This step will take a few seconds.
For European ancestry:
$LASER/laser -g $HGDP/HGDP.633K.euro.geno -c $HGDP/HGDP.633K.euro.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-Euro &
For East Asian ancestry:
$LASER/laser -g $HGDP/HGDP.633K.easia.2.geno -c $HGDP/HGDP.633K.easia.2.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-EAsia &
For Central/South Asian ancestry:
$LASER/laser -g $HGDP/HGDP.633K.csasia.2.geno -c $HGDP/HGDP.633K.csasia.2.RefPC.coord -s $OUT/ancestry/$SAMPLE.HGDP.seq -K 20 -k 4 -M 0.8 -o $OUT/ancestry/$SAMPLE.HGDP-CSAsia &
View the results for European ancestry:
less -S $OUT/ancestry/${SAMPLE}.HGDP-Euro.SeqPC.coord
or for East Asian ancestry
less -S $OUT/ancestry/${SAMPLE}.HGDP-EAsia.SeqPC.coord
or for Central/South Asian ancestry
less -S $OUT/ancestry/${SAMPLE}.HGDP-CSAsia.SeqPC.coord
Visualizing Ancestry
Change to that new directory:
cd $OUT/ancestry/plot
Move your other plot so you don't over-write it
mv Results_on_HGDP.pdf Results_on_HGDP_All.pdf
Generate the plot for European ancestry:
Rscript plotHGDP.r $HGDP/HGDP.633K.euro.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-Euro.SeqPC.coord
or for East Asian ancestry
Rscript plotHGDP.r $HGDP/HGDP.633K.easia.2.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-EAsia.SeqPC.coord
or for Central/South Asian ancestry
Rscript plotHGDP.r $HGDP/HGDP.633K.csasia.2.RefPC.coord $OUT/ancestry/${SAMPLE}.HGDP-CSAsia.SeqPC.coord
Take a look:
evince Results_on_HGDP.pdf &
Return to Calling your own Genome
Go to SeqShop:_Calling_Your_Own_Genome,_December_2014#Reviewing_Indel_Results and pick up where we left off.