Difference between revisions of "SeqShop: Calling Your Own Genome, December 2014"
m (Mktrost moved page SeqShop: Calling Your Own Genome to SeqShop: Calling Your Own Genome, December 2014) |
|||
| (66 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __TOC__ | ||
| + | <div class="mw-collapsible" style="width:500px"> | ||
| + | ''Login instructions for seqshop-server'' | ||
| + | <div class="mw-collapsible-content"> | ||
{{SeqShopLogin}} | {{SeqShopLogin}} | ||
| + | </div> | ||
| + | </div> | ||
| − | == Setup == | + | <div class="mw-collapsible mw-collapsed" style="width:500px"> |
| − | + | == Tuesday - Start INDEL Calling == | |
| + | <div class="mw-collapsible-content"> | ||
| + | === Setup === | ||
| + | The indel & snpcall pipelines will run overnight, but you'll want to log out. | ||
| + | ; How do I leave something running on the server even if I log out? | ||
| + | : One solution is screen! | ||
| − | + | ; How do I use screen? | |
| − | + | : Before running your command, you need to start screen: | |
| − | + | : <pre>screen</pre> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[File:Screen.png]] | |
| − | + | As it says, press <code>Space</code> or <code>Return</code>. | |
| − | + | * It should now look basically the same as your normal command line. | |
| − | = | + | Set these values. Also, be sure to specify your sample name instead of SampleXX |
| + | export SAMPLE=SampleXX | ||
| + | source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | ||
| − | + | See the settings you just used: | |
| − | + | cat /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | |
| + | Shows you: | ||
| + | :<code>export GC=/net/seqshop-server/home/mktrost/seqshop/gotcloud</code> | ||
| + | :<code>export SS=/net/seqshop-server/home/mktrost/seqshop/singleSample</code> | ||
| + | :<code>export OUT=~/$SAMPLE/output</code> | ||
| − | + | === List of BAMs === | |
| − | + | The list of BAMs has already been created (just 1 BAM, your sample). | |
| + | * But it is simply SAMPLE\tBAM_name, so easy to figure out | ||
| − | + | cat ~/$SAMPLE/output/bam.list | |
| − | |||
| − | |||
| − | + | :<code>SampleXX SampleXX/output/bams/SampleXX.recal.bam</code> | |
| − | + | * Relative path, so assumes running from your home directory (I prefer absolute paths, but for simplicity of the workshop, we just use relative path). | |
| − | + | === Configuring Indel === | |
| − | + | No special Configuration settings for Indel calling | |
| + | cat ~/$SAMPLE/gotcloud.conf | ||
| − | + | You will see something like this: | |
| + | <pre> | ||
| + | # Cluster Settings | ||
| + | BATCH_TYPE = | ||
| + | BATCH_OPTS = | ||
| − | + | OUT_DIR = Sample13/output | |
| − | |||
| − | |||
| − | + | # Align Settings | |
| − | + | MAP_TYPE = BWA_MEM | |
| + | BWA_THREADS = -t 24 | ||
| + | FASTQ_LIST = fastq.list | ||
| − | + | # SNP Call Settings | |
| + | UNIT_CHUNK = 20000000 # Chunk size of SNP calling : 20Mb | ||
| + | VCF_EXTRACT = /net/seqshop-server/home/mktrost/seqshop/singleSample/snpOnly.vcf.gz | ||
| + | MODEL_GLFSINGLE = TRUE | ||
| + | MODEL_SKIP_DISCOVER = FALSE | ||
| + | MODEL_AF_PRIOR = TRUE | ||
| − | + | EXT_DIR = /net/seqshop-server/home/mktrost/seqshop/singleSample/ext | |
| − | + | EXT = $(EXT_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(EXT_DIR)/chrCHR.filtered.sites.vcf.gz | |
| + | </pre> | ||
| − | + | === Running Indel Calling === | |
| + | Run GotCloud indel with 6 jobs running in parallel | ||
| + | * Why 6? | ||
| + | ** You want to run as many as you can. | ||
| + | ** 5 of you on the machine - 5*6 = 30 jobs will be running in parallel on that machine | ||
| + | ${GC}/gotcloud indel --conf $SAMPLE/gotcloud.conf --numjobs 6 | ||
| + | * Only need the configuration & number of threads, rest is specified within the configuration. | ||
| − | + | This will run overnight. We will check if it completed at the practical in the morning. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | === Log Out === |
| − | + | ;Want to log out and leave your job running? | |
| + | In the screen window, type: | ||
| + | Ctrl-a d | ||
| + | (Hold down Ctrl and type 'a', let go of both and type 'd') | ||
| + | * This will "detach" from your screen session while your alignment continues to run. | ||
| − | + | If you have not detached from screen: | |
| − | + | Ctrl-a d | |
| − | + | exit PuTTY | |
| − | |||
| − | + | </div> | |
| − | + | </div> | |
| − | + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | |
| − | |||
| − | |||
| − | + | == Wednesday == | |
| − | + | <div class="mw-collapsible-content"> | |
| − | + | <div class="mw-collapsible" style="width:500px"> | |
| − | + | === Checking if INDEL Completed === | |
| − | + | <div class="mw-collapsible-content"> | |
| + | ==== Logging Back in to Check Jobs ==== | ||
| − | + | ;How do you log back into screen? | |
| − | + | screen -r | |
| − | + | This will resume an already running screen. | |
| − | + | Verify you got a "completed successfully" message. | |
| − | # | + | |
| − | + | How long did INDEL calling take? Look at the log message - time in seconds. | |
| + | |||
| + | === Need a BAM Index File? === | ||
| + | Check your BAM files | ||
| + | ls $SAMPLE/output/bams/*bam | ||
| + | |||
| + | Check your BAI files | ||
| + | ls $SAMPLE/output/bams/*bai | ||
| + | |||
| + | Do you have the same number of BAM & BAI files? | ||
| + | * If so, then you are good to go. | ||
| + | * If you have more BAI's than BAM's - oops!!! (LET ME KNOW) | ||
| + | * If you have more BAM's than BAI's, we need to generate the index: | ||
| + | *: <pre>$GC/bin/samtools index $SAMPLE/output/bams/YOUR_BAM_WITH_MISSING_BAI.bam</pre> | ||
| + | |||
| + | |||
| + | === Detach From screen=== | ||
| + | Detach from screen. We will resume it again later when we start SNPCall. | ||
| + | Ctrl-a d | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | === Structural Variation Tutorial === | ||
| + | Now we are going to run the Structural Variation Practical | ||
| + | |||
| + | Please go to: [[SeqShop: Analysis of Structural Variation Practical, December 2014]] | ||
| + | |||
| + | We will Start SNP Calling after the practical. | ||
| + | |||
| + | |||
| + | <div class="mw-collapsible" style="width:500px"> | ||
| + | |||
| + | === Start SNP Calling === | ||
| + | <div class="mw-collapsible-content"> | ||
| + | ==== Resume screen ==== | ||
| + | |||
| + | ;How do you log back into screen? | ||
| + | screen -r | ||
| + | This will resume an already running screen. | ||
| + | |||
| + | Your screen session still has your environment variables set, so you do not need to reset them. | ||
| + | |||
| + | ==== List of BAMs ==== | ||
| + | The list of BAMs has already been created (just 1 BAM, your sample). | ||
| + | * But it is simply SAMPLE\tBAM_name, so easy to figure out | ||
| + | |||
| + | cat ~/$SAMPLE/output/bam.list | ||
| + | |||
| + | :<code>SampleXX SampleXX/output/bams/SampleXX.recal.bam</code> | ||
| + | * Relative path, so assumes running from your home directory (I prefer absolute paths, but for simplicity of the workshop, we just use relative path). | ||
| + | |||
| + | ==== GotCloud SnpCall Configuration ==== | ||
| + | |||
| + | cat ~/$SAMPLE/gotcloud.conf | ||
| + | |||
| + | You will see something like this: | ||
| + | <pre> | ||
| + | # Cluster Settings | ||
| + | BATCH_TYPE = | ||
| + | BATCH_OPTS = | ||
| + | |||
| + | OUT_DIR = Sample13/output | ||
| + | |||
| + | # Align Settings | ||
| + | MAP_TYPE = BWA_MEM | ||
| + | BWA_THREADS = -t 24 | ||
| + | FASTQ_LIST = fastq.list | ||
| + | |||
| + | # SNP Call Settings | ||
| + | UNIT_CHUNK = 20000000 # Chunk size of SNP calling : 20Mb | ||
| + | VCF_EXTRACT = /net/seqshop-server/home/mktrost/seqshop/singleSample/snpOnly.vcf.gz | ||
| + | MODEL_GLFSINGLE = TRUE | ||
| + | MODEL_SKIP_DISCOVER = FALSE | ||
| + | MODEL_AF_PRIOR = TRUE | ||
| + | |||
| + | EXT_DIR = /net/seqshop-server/home/mktrost/seqshop/singleSample/ext | ||
| + | EXT = $(EXT_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(EXT_DIR)/chrCHR.filtered.sites.vcf.gz | ||
| + | </pre> | ||
| + | |||
| + | ===== Configuration Updates ===== | ||
| + | '''No configuration updates from the original settings are necessary.''' | ||
| + | * Originally you were going to update the configuration to do exome calling only, but we have decided to do whole genome | ||
| + | ** If it doesn't finish tonight, we will kill it at the tutorial in the morning, and take advantage of the GotCloud restart capability after the tutorial tomorrow. | ||
| + | |||
| + | If you started making updates to your configuration yesterday, you can try to make it match above (mktrost is actually the path you want in the paths above). | ||
| + | * The only difference from above should be your OUT_DIR. | ||
| + | Check it with (leave as ~mktrost - you are comparing to my NA12878 conf file): | ||
| + | diff $SAMPLE/gotcloud.conf ~mktrost/NA12878/gotcloud.conf | ||
| + | |||
| + | '''If you see differences other than OUT_DIR, you can modify your file using nedit (or your favorite editor as you used yesterday).''' | ||
| + | * '''Or you can copy my file and just change OUT_DIR:''' | ||
| + | cp ~mktrost/NA12878/gotcloud.conf $SAMPLE/gotcloud.conf | ||
| + | nedit $SAMPLE/gotcloud.conf& | ||
| + | * Edit OUT_DIR | ||
| + | OUT_DIR = Sample##/output | ||
| + | * Save, and close | ||
| + | |||
| + | ==== Running SnpCall ==== | ||
| + | Run GotCloud snpcall with 6 jobs running in parallel | ||
| + | * Why 6? | ||
| + | ** You want to run as many as you can. | ||
| + | ** 5 of you on the machine - 5*6 = 30 jobs will be running in parallel on that machine | ||
| + | ${GC}/gotcloud snpcall --conf $SAMPLE/gotcloud.conf --numjobs 6 | ||
| + | * Only need the configuration & number of threads, rest is specified within the configuration. | ||
| + | |||
| + | This will run overnight. We will check if it completed at the practical in the morning. | ||
| + | |||
| + | ==== Log Out ==== | ||
| + | ;Want to log out and leave your job running? | ||
| + | In the screen window, type: | ||
| + | Ctrl-a d | ||
| + | (Hold down Ctrl and type 'a', let go of both and type 'd') | ||
| + | * This will "detach" from your screen session while your alignment continues to run. | ||
| + | |||
| + | exit PuTTY | ||
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | ||
| + | |||
| + | == Thursday == | ||
| + | <div class="mw-collapsible-content"> | ||
| + | === Checking if SnpCall Completed === | ||
| + | ==== Logging Back in to Check Jobs ==== | ||
| + | |||
| + | ;How do you log back into screen? | ||
| + | screen -r | ||
| + | This will resume an already running screen. | ||
| + | |||
| + | ==== Checking Completion ==== | ||
| + | |||
| + | Did you get a "completed successfully" message? | ||
| + | |||
| + | If yes, how long did SNP calling take? Look at the log message - time in seconds. | ||
| + | |||
| + | If no, are you running on seqshop-server? If so, KILL it. ssh -X to one of the seqshop machines and run on there. | ||
| + | Ctrl-c | ||
| + | |||
| + | Detach from screen. We will resume it again later when we restart SNPCall (if necessary). | ||
| + | Ctrl-a d | ||
| + | |||
| + | === Ancestry Tutorial === | ||
| + | Now we are going to run the Structural Variation Practical | ||
| + | |||
| + | Please go to: [[SeqShop: Estimates of Genetic Ancestry Practical, December 2014]] | ||
| + | |||
| + | We will Resume SNP Calling (if necessary) after the practical. | ||
| + | |||
| + | |||
| + | === Restart SnpCall === | ||
| + | ==== Resume screen ==== | ||
| + | |||
| + | ;How do you log back into screen? | ||
| + | screen -r | ||
| + | This will resume an already running screen. | ||
| + | |||
| + | Your screen session still has your environment variables set, so you do not need to reset them. | ||
| + | |||
| + | ==== Running SnpCall ==== | ||
| + | Run GotCloud snpcall with 6 jobs running in parallel | ||
| + | ${GC}/gotcloud snpcall --conf $SAMPLE/gotcloud.conf --numjobs 6 | ||
| + | * GotCloud will pick up after the last completed step from before. | ||
| + | |||
| + | This will run overnight. We will check if it completed at the practical in the morning. | ||
| + | |||
| + | ==== Log Out ==== | ||
| + | ;Want to log out and leave your job running? | ||
| + | In the screen window, type: | ||
| + | Ctrl-a d | ||
| + | (Hold down Ctrl and type 'a', let go of both and type 'd') | ||
| + | * This will "detach" from your screen session while your alignment continues to run. | ||
| + | |||
| + | exit PuTTY | ||
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="mw-collapsible" style="width:1000px"> | ||
| + | |||
| + | == Friday == | ||
| + | <div class="mw-collapsible-content"> | ||
| + | |||
| + | |||
| + | === SeqShop: Ancestry On Your Own Genome, December 2014 === | ||
| + | [[SeqShop: Ancestry On Your Own Genome, December 2014]] | ||
| + | |||
| + | |||
| + | === Association Analysis Tutorial === | ||
| + | Now we are going to run the Association Analysis Practical | ||
| + | |||
| + | Please go to: [[SeqShop: Genetic Association Analysis Practical, December 2014]] | ||
| + | |||
| + | We will look at our own genomes again after the practical. | ||
| + | |||
| + | |||
| + | === Return to SeqShop: Ancestry On Your Own Genome, December 2014 === | ||
| + | Return to [[SeqShop:_Ancestry_On_Your_Own_Genome,_December_2014#Checking_if_Pileup_finished]] | ||
| + | |||
| + | === Reviewing Indel Results === | ||
| + | Set these values. Also, be sure to specify your sample name (or NA12878) instead of SampleXX | ||
| + | export SAMPLE=SampleXX | ||
| + | source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | ||
| + | |||
| + | Look in the output directory | ||
| + | ls ~/$SAMPLE/output | ||
| + | |||
| + | What in that directory was produced by indel calling? | ||
| + | * <code>gotcloud.indel.conf</code> | ||
| + | ** dump of all configuration settings for this run | ||
| + | * <code>gotcloud.indel.Makefile</code> | ||
| + | ** Makefile that was generated to manage all of the commands to be run | ||
| + | * <code>gotcloud.indel.Makefile.log</code> | ||
| + | ** log of all commands run by the Makefile | ||
| + | * <code>indel/</code> | ||
| + | ** indel output directory | ||
| + | |||
| + | Let's look at the indel output | ||
| + | ls ~/$SAMPLE/output/indel | ||
| + | * 3 directories | ||
| + | ** aux - intermediate files | ||
| + | ** indelvcf - intermediate files | ||
| + | ** '''final indel files''' | ||
| + | |||
| + | Final indel directory: | ||
| + | ls ~/$SAMPLE/output/indel/final | ||
| + | * '''all.genotypes.vcf.gz - output VCF''' | ||
| + | * '''all.genotypes.vcf.gz.tbi - output VCF index file to allow jumping to positions''' | ||
| + | * all.genotypes.vcf.gz.tbi.OK - completion indicator | ||
| + | * merge/ - directory with per chromosome bcf (binary vcf) files | ||
| + | * all.genotypes.vcf.gz.OK - completion indicator | ||
| + | * all.genotypes.vcf.gz.tbi.log - log | ||
| + | * concat.log - log | ||
| + | |||
| + | ==== Looking at final INDEL VCF ==== | ||
| + | |||
| + | Note that because this is a single sample calling, many of the INFO fields are less meaningful as many of the values like HWE p values, allele frequencies, inbreeding coefficient are a function of a population. | ||
| + | Nonetheless, we may examine the results. First, we see how many indels were discovered for your genome: | ||
| + | |||
| + | $GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz | ||
| + | |||
| + | no. Indels : 588566 | ||
| + | 2 alleles (ins/del) : 588566 (0.87) [273261/315305] | ||
| + | |||
| + | This gives use 588,566 indels with an insertion deletion ratio of 0.87. | ||
| + | |||
| + | We next look at the filtered set. The PASS filter reduces the setof indels to a non overlapping set and the INFO.AC!=0 extracts all indels that are either heterozygous or homozygous alternative. | ||
| + | Some indels that were originally discovered were found to be the homozygous reference genotype. Invariably, these are relative high depth calls where the | ||
| + | alternative allele is discovered less or is mis-specified. | ||
| + | |||
| + | $GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz -f "PASS&&INFO.AC!=0" | ||
| + | |||
| + | no. Indels : 549963 | ||
| + | 2 alleles (ins/del) : 549963 (0.91) [261480/288483] | ||
| + | |||
| + | About 38K indels were removed, the insertion deletion ratio increases to 0.91. Note that in general, for high depth data, discovered indels are reported with insertion deletion ratios | ||
| + | close to 1. So this is a good sign. Next generation sequencing errors are bias for deletions. | ||
| + | |||
| + | It is possible to perform a slightly more stringent filtering using allele balance. The allele balance estimator in this case is meaningful still for an individual because it is a function of read depth. | ||
| + | Note that AB>0.5 denotes reference bias and AB<0.5 denotes alternative allele bias. | ||
| + | |||
| + | $GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz -f "PASS&&INFO.AC>0&&INFO.AB<0.7&&INFO.AB>0.3" | ||
| + | |||
| + | no. Indels : 490965 | ||
| + | 2 alleles (ins/del) : 490965 (0.92) [235254/255711] | ||
| + | |||
| + | The insertion deletion ratio increases from 0.91 to 0.92. | ||
| + | |||
| + | |||
| + | === Friday: Reviewing SNPCALL Results === | ||
| + | Look in the output directory | ||
| + | ls ~/$SAMPLE/output | ||
| + | |||
| + | Look at the vcfs: | ||
| + | ls ~/$SAMPLE/output/vcfs | ||
| + | |||
| + | === Friday : More SNP Analysis === | ||
| + | |||
| + | ==== Environmental Variables ==== | ||
| + | |||
| + | If you didn't set the environmental variable, you can set it again | ||
| + | |||
| + | source /net/seqshop-server/home/mktrost/seqshop/setup.txt | ||
| + | export SAMPLE=SampleXX (MAKE SURE TO CHANGE XX to your number or use NA12878 instead) | ||
| + | source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt | ||
| + | |||
| + | In addition, set another environmental variable for locating the binaries for custom analysis | ||
| + | |||
| + | export HK=/net/seqshop-server/home/hmkang/seqshop/bin | ||
| + | |||
| + | ==== Annotation / Lookup against dbSNP ==== | ||
| + | |||
| + | If you want to add rsIDs to your variant files, you can do this by running the following command | ||
| + | |||
| + | $HK/vcf-add-rsid -vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --db $HK/../data/dbSNP.b138/dbsnp_138.b37.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz | ||
| + | |||
| + | If you want to run this command across all chromosomes in parallel, you can use the special script run-command-wgs | ||
| + | |||
| + | $HK/run-command-wgs --cmd "$HK/vcf-add-rsid -vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --db $HK/../data/dbSNP.b138/dbsnp_138.b37.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz" --numjobs 6 | ||
| + | |||
| + | Looking up SNPs by rsID is possible by (for example) | ||
| + | $HK/vcf-lookup-rsid --vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --sepchr --rs rs17766217 | ||
| + | * Be sure to look at the QUAL & your sample's PL, and not just the GL field. Check if QUAL is 0 or PL is 0,0,0 - NS is also probably 0; DP is probably 0. That means you probably didn't have any copies, so your GT may not be correct/is unknown. | ||
| + | |||
| + | If you want to browse the rsIDs of known GWAS SNPs, you can do this by | ||
| + | cut -f 1,8,22 $HK/../data/gwascatalog/gwascatalog.txt | less | ||
| + | |||
| + | ==== Annotating your genome ==== | ||
| + | |||
| + | You can annotate your genome using EPACTS software packages. Individual chromosome can be annotated by running. | ||
| + | $EPACTS/bin/epacts anno --in $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz | ||
| + | |||
| + | Or you can run multiple chromosomes in parallel in one command | ||
| + | $HK/run-command-wgs --cmd "$EPACTS/bin/epacts anno --in $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz" --numjobs 6 | ||
| + | |||
| + | ==== Extracting only exonic SNPs ==== | ||
| + | |||
| + | If you want to look at the exonic SNPs, you can extract using the following command | ||
| + | $HK/run-command-wgs --cmd "($HK/tabix -H $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz; zcat $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz | grep Exon;)| $HK/bgzip -c > $OUT/vcfs/chr1/chr1.filtered.rsid.anno.exon.vcf.gz" --numjobs 6 | ||
| + | |||
| + | And they can be combined as follows | ||
| + | (zcat $OUT/vcfs/chr1/chr1.filtered.rsid.anno.exon.vcf.gz; zcat $OUT/vcfs/chr[2-9]/chr*.filtered.rsid.anno.exon.vcf.gz $OUT/vcfs/chr??/chr*.filtered.rsid.anno.exon.vcf.gz $OUT/vcfs/chrX/chrX.filtered.rsid.anno.exon.vcf.gz | grep -v ^#) | $HK/bgzip -c > $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | ||
| + | |||
| + | ==== Exonic Variants NOT found by 1000G ==== | ||
| + | |||
| + | If you are interested in rare variants that are not identified by 1000G, you can extract them by running | ||
| + | zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | less | ||
| − | + | For example, | |
| − | + | zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | perl -lane 'print "$1\t$F[6]" if ( /ANNO=([^;:]+)/)' | sort | uniq -c | |
| − | + | will give you the counts of variants, separate by the filtering results | |
| − | + | ||
| + | * Q1. How manny novel silent, missense, and nonsense SNPs are found? Is that too few, too small, or just about right? | ||
| + | * Q2. Looking at each functional category, which functional categories has largest fraction of SNPs failed filter? Why do you think it is? | ||
| + | * Q3. Can you exclude the sites that are also in dbSNP, and count how many nonsense variants are left? | ||
| + | |||
| + | |||
| + | To also exclude those in dbsnp: | ||
| + | zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | grep -v rs| perl -lane 'print "$1\t$F[6]" if ( /ANNO=([^;:]+)/)' | sort | uniq -c | ||
| − | + | Exclude dbsnp and look at Stop_Gain variants | |
| + | zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" |grep -v rs | perl -lane 'print "$_" if ( /ANNO=Stop_Gain/)' |grep -w PASS | ||
| + | |||
| + | Want to see this from the BAM file? Use samtools tview: | ||
| + | $GC/bin/samtools tview $SAMPLE/output/bams/$SAMPLE.recal.bam $GC/gotcloud.ref/human.g1k.v37.fa | ||
| + | Use 'g' & enter the Chr:Pos | ||
| + | * Some patterns may indicate not real variants. | ||
| + | |||
| + | If you want to know predicted functional significance of a particular variant, you can search by | ||
| + | |||
| + | $HK/tabix $HK/../data/CADD/whole_genome_SNVs.tsv.gz [chr]:[pos] | head -3 | ||
| + | |||
| + | The phred score at the last column quantifies the degree of functional significance | ||
| − | |||
| − | |||
| − | |||
| − | + | </div> | |
| − | + | </div> | |
| − | |||
| − | |||
| − | |||
Latest revision as of 12:16, 15 December 2014
Login instructions for seqshop-server
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
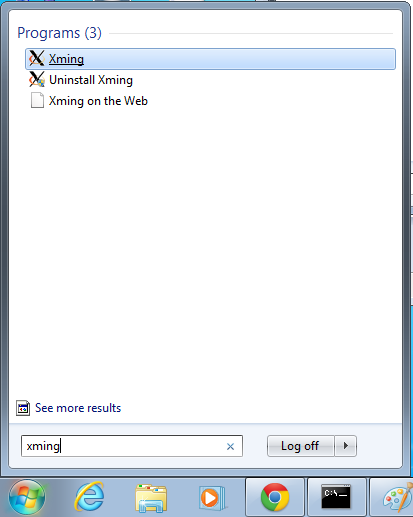
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
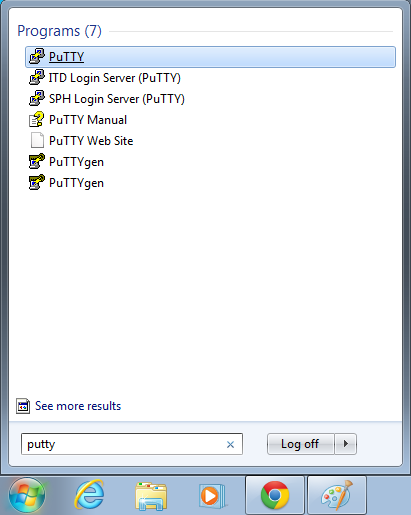
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
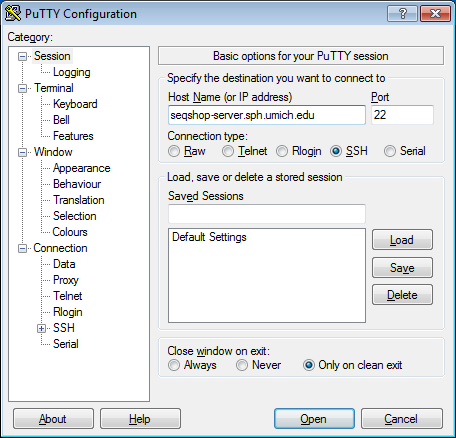
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
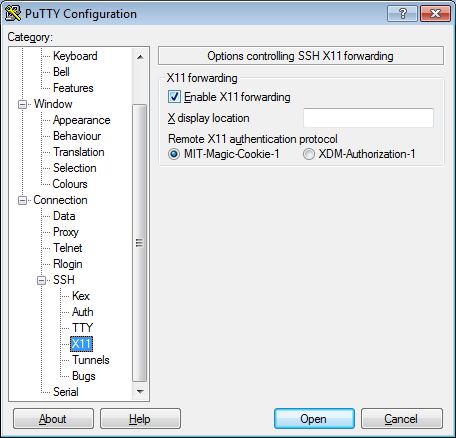
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Tuesday - Start INDEL Calling
Setup
The indel & snpcall pipelines will run overnight, but you'll want to log out.
- How do I leave something running on the server even if I log out?
- One solution is screen!
- How do I use screen?
- Before running your command, you need to start screen:
screen
As it says, press Space or Return.
- It should now look basically the same as your normal command line.
Set these values. Also, be sure to specify your sample name instead of SampleXX
export SAMPLE=SampleXX source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
See the settings you just used:
cat /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
Shows you:
export GC=/net/seqshop-server/home/mktrost/seqshop/gotcloudexport SS=/net/seqshop-server/home/mktrost/seqshop/singleSampleexport OUT=~/$SAMPLE/output
List of BAMs
The list of BAMs has already been created (just 1 BAM, your sample).
- But it is simply SAMPLE\tBAM_name, so easy to figure out
cat ~/$SAMPLE/output/bam.list
SampleXX SampleXX/output/bams/SampleXX.recal.bam
- Relative path, so assumes running from your home directory (I prefer absolute paths, but for simplicity of the workshop, we just use relative path).
Configuring Indel
No special Configuration settings for Indel calling
cat ~/$SAMPLE/gotcloud.conf
You will see something like this:
# Cluster Settings BATCH_TYPE = BATCH_OPTS = OUT_DIR = Sample13/output # Align Settings MAP_TYPE = BWA_MEM BWA_THREADS = -t 24 FASTQ_LIST = fastq.list # SNP Call Settings UNIT_CHUNK = 20000000 # Chunk size of SNP calling : 20Mb VCF_EXTRACT = /net/seqshop-server/home/mktrost/seqshop/singleSample/snpOnly.vcf.gz MODEL_GLFSINGLE = TRUE MODEL_SKIP_DISCOVER = FALSE MODEL_AF_PRIOR = TRUE EXT_DIR = /net/seqshop-server/home/mktrost/seqshop/singleSample/ext EXT = $(EXT_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(EXT_DIR)/chrCHR.filtered.sites.vcf.gz
Running Indel Calling
Run GotCloud indel with 6 jobs running in parallel
- Why 6?
- You want to run as many as you can.
- 5 of you on the machine - 5*6 = 30 jobs will be running in parallel on that machine
${GC}/gotcloud indel --conf $SAMPLE/gotcloud.conf --numjobs 6
- Only need the configuration & number of threads, rest is specified within the configuration.
This will run overnight. We will check if it completed at the practical in the morning.
Log Out
- Want to log out and leave your job running?
In the screen window, type:
Ctrl-a d
(Hold down Ctrl and type 'a', let go of both and type 'd')
- This will "detach" from your screen session while your alignment continues to run.
If you have not detached from screen:
Ctrl-a d
exit PuTTY
Wednesday
Checking if INDEL Completed
Logging Back in to Check Jobs
- How do you log back into screen?
screen -r
This will resume an already running screen.
Verify you got a "completed successfully" message.
How long did INDEL calling take? Look at the log message - time in seconds.
Need a BAM Index File?
Check your BAM files
ls $SAMPLE/output/bams/*bam
Check your BAI files
ls $SAMPLE/output/bams/*bai
Do you have the same number of BAM & BAI files?
- If so, then you are good to go.
- If you have more BAI's than BAM's - oops!!! (LET ME KNOW)
- If you have more BAM's than BAI's, we need to generate the index:
$GC/bin/samtools index $SAMPLE/output/bams/YOUR_BAM_WITH_MISSING_BAI.bam
Detach From screen
Detach from screen. We will resume it again later when we start SNPCall.
Ctrl-a d
Structural Variation Tutorial
Now we are going to run the Structural Variation Practical
Please go to: SeqShop: Analysis of Structural Variation Practical, December 2014
We will Start SNP Calling after the practical.
Start SNP Calling
Resume screen
- How do you log back into screen?
screen -r
This will resume an already running screen.
Your screen session still has your environment variables set, so you do not need to reset them.
List of BAMs
The list of BAMs has already been created (just 1 BAM, your sample).
- But it is simply SAMPLE\tBAM_name, so easy to figure out
cat ~/$SAMPLE/output/bam.list
SampleXX SampleXX/output/bams/SampleXX.recal.bam
- Relative path, so assumes running from your home directory (I prefer absolute paths, but for simplicity of the workshop, we just use relative path).
GotCloud SnpCall Configuration
cat ~/$SAMPLE/gotcloud.conf
You will see something like this:
# Cluster Settings BATCH_TYPE = BATCH_OPTS = OUT_DIR = Sample13/output # Align Settings MAP_TYPE = BWA_MEM BWA_THREADS = -t 24 FASTQ_LIST = fastq.list # SNP Call Settings UNIT_CHUNK = 20000000 # Chunk size of SNP calling : 20Mb VCF_EXTRACT = /net/seqshop-server/home/mktrost/seqshop/singleSample/snpOnly.vcf.gz MODEL_GLFSINGLE = TRUE MODEL_SKIP_DISCOVER = FALSE MODEL_AF_PRIOR = TRUE EXT_DIR = /net/seqshop-server/home/mktrost/seqshop/singleSample/ext EXT = $(EXT_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(EXT_DIR)/chrCHR.filtered.sites.vcf.gz
Configuration Updates
No configuration updates from the original settings are necessary.
- Originally you were going to update the configuration to do exome calling only, but we have decided to do whole genome
- If it doesn't finish tonight, we will kill it at the tutorial in the morning, and take advantage of the GotCloud restart capability after the tutorial tomorrow.
If you started making updates to your configuration yesterday, you can try to make it match above (mktrost is actually the path you want in the paths above).
- The only difference from above should be your OUT_DIR.
Check it with (leave as ~mktrost - you are comparing to my NA12878 conf file):
diff $SAMPLE/gotcloud.conf ~mktrost/NA12878/gotcloud.conf
If you see differences other than OUT_DIR, you can modify your file using nedit (or your favorite editor as you used yesterday).
- Or you can copy my file and just change OUT_DIR:
cp ~mktrost/NA12878/gotcloud.conf $SAMPLE/gotcloud.conf nedit $SAMPLE/gotcloud.conf&
- Edit OUT_DIR
OUT_DIR = Sample##/output
- Save, and close
Running SnpCall
Run GotCloud snpcall with 6 jobs running in parallel
- Why 6?
- You want to run as many as you can.
- 5 of you on the machine - 5*6 = 30 jobs will be running in parallel on that machine
${GC}/gotcloud snpcall --conf $SAMPLE/gotcloud.conf --numjobs 6
- Only need the configuration & number of threads, rest is specified within the configuration.
This will run overnight. We will check if it completed at the practical in the morning.
Log Out
- Want to log out and leave your job running?
In the screen window, type:
Ctrl-a d
(Hold down Ctrl and type 'a', let go of both and type 'd')
- This will "detach" from your screen session while your alignment continues to run.
exit PuTTY
Thursday
Checking if SnpCall Completed
Logging Back in to Check Jobs
- How do you log back into screen?
screen -r
This will resume an already running screen.
Checking Completion
Did you get a "completed successfully" message?
If yes, how long did SNP calling take? Look at the log message - time in seconds.
If no, are you running on seqshop-server? If so, KILL it. ssh -X to one of the seqshop machines and run on there.
Ctrl-c
Detach from screen. We will resume it again later when we restart SNPCall (if necessary).
Ctrl-a d
Ancestry Tutorial
Now we are going to run the Structural Variation Practical
Please go to: SeqShop: Estimates of Genetic Ancestry Practical, December 2014
We will Resume SNP Calling (if necessary) after the practical.
Restart SnpCall
Resume screen
- How do you log back into screen?
screen -r
This will resume an already running screen.
Your screen session still has your environment variables set, so you do not need to reset them.
Running SnpCall
Run GotCloud snpcall with 6 jobs running in parallel
${GC}/gotcloud snpcall --conf $SAMPLE/gotcloud.conf --numjobs 6
- GotCloud will pick up after the last completed step from before.
This will run overnight. We will check if it completed at the practical in the morning.
Log Out
- Want to log out and leave your job running?
In the screen window, type:
Ctrl-a d
(Hold down Ctrl and type 'a', let go of both and type 'd')
- This will "detach" from your screen session while your alignment continues to run.
exit PuTTY
Friday
SeqShop: Ancestry On Your Own Genome, December 2014
SeqShop: Ancestry On Your Own Genome, December 2014
Association Analysis Tutorial
Now we are going to run the Association Analysis Practical
Please go to: SeqShop: Genetic Association Analysis Practical, December 2014
We will look at our own genomes again after the practical.
Return to SeqShop: Ancestry On Your Own Genome, December 2014
Return to SeqShop:_Ancestry_On_Your_Own_Genome,_December_2014#Checking_if_Pileup_finished
Reviewing Indel Results
Set these values. Also, be sure to specify your sample name (or NA12878) instead of SampleXX
export SAMPLE=SampleXX source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
Look in the output directory
ls ~/$SAMPLE/output
What in that directory was produced by indel calling?
gotcloud.indel.conf- dump of all configuration settings for this run
gotcloud.indel.Makefile- Makefile that was generated to manage all of the commands to be run
gotcloud.indel.Makefile.log- log of all commands run by the Makefile
indel/- indel output directory
Let's look at the indel output
ls ~/$SAMPLE/output/indel
- 3 directories
- aux - intermediate files
- indelvcf - intermediate files
- final indel files
Final indel directory:
ls ~/$SAMPLE/output/indel/final
- all.genotypes.vcf.gz - output VCF
- all.genotypes.vcf.gz.tbi - output VCF index file to allow jumping to positions
- all.genotypes.vcf.gz.tbi.OK - completion indicator
- merge/ - directory with per chromosome bcf (binary vcf) files
- all.genotypes.vcf.gz.OK - completion indicator
- all.genotypes.vcf.gz.tbi.log - log
- concat.log - log
Looking at final INDEL VCF
Note that because this is a single sample calling, many of the INFO fields are less meaningful as many of the values like HWE p values, allele frequencies, inbreeding coefficient are a function of a population. Nonetheless, we may examine the results. First, we see how many indels were discovered for your genome:
$GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz
no. Indels : 588566
2 alleles (ins/del) : 588566 (0.87) [273261/315305]
This gives use 588,566 indels with an insertion deletion ratio of 0.87.
We next look at the filtered set. The PASS filter reduces the setof indels to a non overlapping set and the INFO.AC!=0 extracts all indels that are either heterozygous or homozygous alternative. Some indels that were originally discovered were found to be the homozygous reference genotype. Invariably, these are relative high depth calls where the alternative allele is discovered less or is mis-specified.
$GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz -f "PASS&&INFO.AC!=0"
no. Indels : 549963
2 alleles (ins/del) : 549963 (0.91) [261480/288483]
About 38K indels were removed, the insertion deletion ratio increases to 0.91. Note that in general, for high depth data, discovered indels are reported with insertion deletion ratios close to 1. So this is a good sign. Next generation sequencing errors are bias for deletions.
It is possible to perform a slightly more stringent filtering using allele balance. The allele balance estimator in this case is meaningful still for an individual because it is a function of read depth. Note that AB>0.5 denotes reference bias and AB<0.5 denotes alternative allele bias.
$GC/bin/vt peek ~/$SAMPLE/output/indel/final/all.genotypes.vcf.gz -f "PASS&&INFO.AC>0&&INFO.AB<0.7&&INFO.AB>0.3"
no. Indels : 490965
2 alleles (ins/del) : 490965 (0.92) [235254/255711]
The insertion deletion ratio increases from 0.91 to 0.92.
Friday: Reviewing SNPCALL Results
Look in the output directory
ls ~/$SAMPLE/output
Look at the vcfs:
ls ~/$SAMPLE/output/vcfs
Friday : More SNP Analysis
Environmental Variables
If you didn't set the environmental variable, you can set it again
source /net/seqshop-server/home/mktrost/seqshop/setup.txt export SAMPLE=SampleXX (MAKE SURE TO CHANGE XX to your number or use NA12878 instead) source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
In addition, set another environmental variable for locating the binaries for custom analysis
export HK=/net/seqshop-server/home/hmkang/seqshop/bin
Annotation / Lookup against dbSNP
If you want to add rsIDs to your variant files, you can do this by running the following command
$HK/vcf-add-rsid -vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --db $HK/../data/dbSNP.b138/dbsnp_138.b37.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz
If you want to run this command across all chromosomes in parallel, you can use the special script run-command-wgs
$HK/run-command-wgs --cmd "$HK/vcf-add-rsid -vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --db $HK/../data/dbSNP.b138/dbsnp_138.b37.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz" --numjobs 6
Looking up SNPs by rsID is possible by (for example)
$HK/vcf-lookup-rsid --vcf $OUT/vcfs/chr1/chr1.filtered.vcf.gz --sepchr --rs rs17766217
- Be sure to look at the QUAL & your sample's PL, and not just the GL field. Check if QUAL is 0 or PL is 0,0,0 - NS is also probably 0; DP is probably 0. That means you probably didn't have any copies, so your GT may not be correct/is unknown.
If you want to browse the rsIDs of known GWAS SNPs, you can do this by
cut -f 1,8,22 $HK/../data/gwascatalog/gwascatalog.txt | less
Annotating your genome
You can annotate your genome using EPACTS software packages. Individual chromosome can be annotated by running.
$EPACTS/bin/epacts anno --in $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz
Or you can run multiple chromosomes in parallel in one command
$HK/run-command-wgs --cmd "$EPACTS/bin/epacts anno --in $OUT/vcfs/chr1/chr1.filtered.rsid.vcf.gz --out $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz" --numjobs 6
Extracting only exonic SNPs
If you want to look at the exonic SNPs, you can extract using the following command
$HK/run-command-wgs --cmd "($HK/tabix -H $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz; zcat $OUT/vcfs/chr1/chr1.filtered.rsid.anno.vcf.gz | grep Exon;)| $HK/bgzip -c > $OUT/vcfs/chr1/chr1.filtered.rsid.anno.exon.vcf.gz" --numjobs 6
And they can be combined as follows
(zcat $OUT/vcfs/chr1/chr1.filtered.rsid.anno.exon.vcf.gz; zcat $OUT/vcfs/chr[2-9]/chr*.filtered.rsid.anno.exon.vcf.gz $OUT/vcfs/chr??/chr*.filtered.rsid.anno.exon.vcf.gz $OUT/vcfs/chrX/chrX.filtered.rsid.anno.exon.vcf.gz | grep -v ^#) | $HK/bgzip -c > $OUT/wgs.filtered.rsid.anno.exon.vcf.gz
Exonic Variants NOT found by 1000G
If you are interested in rare variants that are not identified by 1000G, you can extract them by running
zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | less
For example,
zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | perl -lane 'print "$1\t$F[6]" if ( /ANNO=([^;:]+)/)' | sort | uniq -c
will give you the counts of variants, separate by the filtering results
- Q1. How manny novel silent, missense, and nonsense SNPs are found? Is that too few, too small, or just about right?
- Q2. Looking at each functional category, which functional categories has largest fraction of SNPs failed filter? Why do you think it is?
- Q3. Can you exclude the sites that are also in dbSNP, and count how many nonsense variants are left?
To also exclude those in dbsnp:
zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" | grep -v rs| perl -lane 'print "$1\t$F[6]" if ( /ANNO=([^;:]+)/)' | sort | uniq -c
Exclude dbsnp and look at Stop_Gain variants
zcat $OUT/wgs.filtered.rsid.anno.exon.vcf.gz | grep "EXTFILTER=NA,NA" | grep -v -w "0/0" |grep -v rs | perl -lane 'print "$_" if ( /ANNO=Stop_Gain/)' |grep -w PASS
Want to see this from the BAM file? Use samtools tview:
$GC/bin/samtools tview $SAMPLE/output/bams/$SAMPLE.recal.bam $GC/gotcloud.ref/human.g1k.v37.fa
Use 'g' & enter the Chr:Pos
- Some patterns may indicate not real variants.
If you want to know predicted functional significance of a particular variant, you can search by
$HK/tabix $HK/../data/CADD/whole_genome_SNVs.tsv.gz [chr]:[pos] | head -3
The phred score at the last column quantifies the degree of functional significance