SeqShop: Estimates of Genetic Ancestry Practical, June 2014
Introduction
See the tutorial slides for an introduction of the LASER analysis workflow, input/output file formats, and usage of the LASER software.
The main purpose of this page is to provide step-by-step command lines for using LASER to estimate ancestry of 6 targeted sequenced samples (2 HapMap trios) in a principal component space generated using genome-wide SNP data from the Human Genome Diversity Project (HGDP).
For more details about the options and usage of LASER, please read the manual.
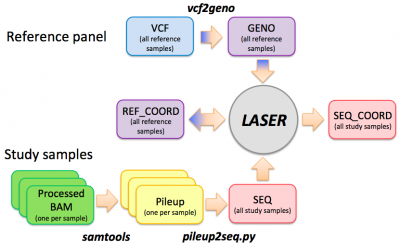
LASER workflow
Getting started
Create a working directory:
mkdir ancestry cd ancestry
Download and decompress software package:
wget http://www.sph.umich.edu/csg/chaolong/LASER/LASER-2.01.tar.gz tar xzvf LASER-2.01.tar.gz
Set up to access data:
source /home/chaolong/LASER-Tutorial/setup.txt
What is in the setup.txt file:
export GC=/home/mktrost/seqshop/gotcloud export BAM=/home/chaolong/LASER-Tutorial/BAM export REF=/home/chaolong/LASER-Tutorial/reference export HGDP=/home/chaolong/LASER-Tutorial/HGDP
Preparing input files for LASER
Step 0: vcf --> geno
This step prepares the reference panel by converting a VCF genotype file to a GENO file. We will skip this step and use a ready-to-use HGDP reference panel. A typical command to run the vcf2geno tool is given in the file "./LASER-2.01/vcf2geno/cmd.sh":
# ./vcf2geno --inVcf exampleVCF/example.vcf.gz --updateID test.updateId --out test
Step 1: bam --> pileup
This step uses samtools to generate pileup files from bam files. Please only try one sample so that we won't overload the sever with everyone running 6 jobs at the same time. Pileup files for these 6 samples have been prepared for later steps. It takes about 2 mins for each pileup job.
$GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101035.recal.bam > 121101035.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101043.recal.bam > 121101043.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101050.recal.bam > 121101050.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101052.recal.bam > 121101052.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101415.recal.bam > 121101415.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/hs37d5.fa.rz -l $HGDP/HGDP_938.bed $BAM/121101861.recal.bam > 121101861.recal.pileup &
Step 2: pileup --> seq
In this step, we will generate a file called "hapmap_trios.seq", containing the information of 6 samples. It takes about 30 seconds to run. We will use the pre-generated pileup files in the $BAM folder.
python ./LASER-2.01/pileup2seq/pileup2seq.py \ -m $HGDP/HGDP_938.site \ -b $BAM/AMD_roi_1-based.bed \ -i $BAM/AMD_hapmap_trios_id.txt \ -o hapmap_trios \ $BAM/121101035.recal.pileup \ $BAM/121101043.recal.pileup \ $BAM/121101050.recal.pileup \ $BAM/121101052.recal.pileup \ $BAM/121101415.recal.pileup \ $BAM/121101861.recal.pileup &
In the above command, -i specifies alternative IDs for the BAM files to be used in the .seq file (including popID and indivID). -b and -i are optional.
Estimating ancestry coordinates using LASER
Step 0: Generate the reference ancestry space (using the PCA mode of the LASER program)
# ./LASER-2.01/laser -g ./HGDP/HGDP_938.geno -pca 1 -k 30 -o HGDP_938
The above command takes about 15 mins to finish. We will skip this step, and use a set of reference ancestry coordinates that have been generated in the file $HGDP/HGDP_938.RefPC.coord. View the reference coordinates:
less -S $HGDP/HGDP_938.RefPC.coord
Step 1: Place sequenced samples into the reference ancestry space:
./LASER-2.01/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s hapmap_trios.seq -K 20 -k 4 -x 1 -y 3 -o hapmap_trios.1-3 & ./LASER-2.01/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s hapmap_trios.seq -K 20 -k 4 -x 4 -y 6 -o hapmap_trios.4-6 &
The first job will process samples 1 to 3 and the second job will processed samples 4 to 6. Each sequenced sample will be projected from a 20-dimensional PCA space onto a 4-dimensional reference ancestry space. The running time is ~10 minutes for processing 3 samples in each job.
Step 2: Combine results
Results from previous step will be output to two files "hapmap_trios.1-3.SeqPC.coord" and "hapmap_trios.4-6.SeqPC.coord". Here we simply concatenate the two files while skipping the header line of the second file.
cp hapmap_trios.1-3.SeqPC.coord hapmap_trios.SeqPC.coord more +2 hapmap_trios.4-6.SeqPC.coord >> hapmap_trios.SeqPC.coord
Step 3: Plot results using R
Example R codes are available in ./LASER-2.01/plot/. Let's first copy the folder to current working directory:
cp -r ./LASER-2.01/plot/ ./
And then we enter the directory and run the script to plot results:
cd ./plot Rscript plotHGDP.r $HGDP/HGDP_938.RefPC.coord ../hapmap_trios.SeqPC.coord
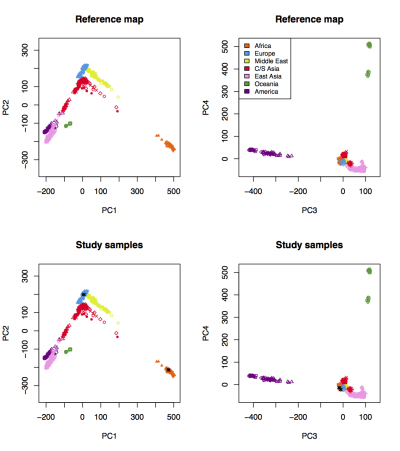
A figure named "Results_on_HGDP.pdf" will be generated. Visualize the figure:
evince Results_on_HGDP.pdf &
We expect to see the following figure: