SeqShop: Estimates of Genetic Ancestry Practical, June 2014
Introduction
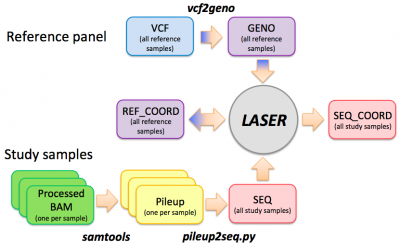
See the tutorial slides for an introduction of the LASER analysis workflow, input/output file formats, and usage of the LASER software.
The main purpose of this page is to provide step-by-step command lines for using LASER to estimate ancestry of 6 targeted sequenced samples (2 HapMap trios) in a principal component space generated using genome-wide SNP data from the Human Genome Diversity Project (HGDP). The HGDP reference panel contains genotype data across 632,958 autosomal loci for 938 individuals from 53 populations worldwide.
For more details about the options and usage of LASER, please read the manual.
LASER workflow
Getting started
Create a working directory:
mkdir ancestry cd ancestry
Download and decompress software package:
wget http://www.sph.umich.edu/csg/chaolong/LASER/LASER-2.01.tar.gz tar xzvf LASER-2.01.tar.gz
Set up to access data:
source /home/chaolong/LASER-Tutorial/setup.txt
What is in the setup.txt file:
export GC=/home/mktrost/seqshop/gotcloud export REF=/home/mktrost/seqshop/reference/all export HGDP=/home/chaolong/LASER-Tutorial/HGDP export BAM=/home/chaolong/LASER-Tutorial/BAM
Preparing input files for LASER
Step 0: vcf --> geno
This step prepares the reference panel by converting a VCF genotype file to a GENO file. We will skip this step and use a ready-to-use HGDP reference panel. A typical command to run the vcf2geno tool is given in the file "./LASER-2.01/vcf2geno/cmd.sh":
# cd ./LASER-2.01/vcf2geno/ # ./vcf2geno --inVcf exampleVCF/example.vcf.gz --updateID test.updateId --out test
Step 1: bam --> pileup
This step uses samtools to generate pileup files from bam files. Please only try one sample so that we won't overload the sever with everyone running 6 jobs at the same time. Pileup files for these 6 samples have been prepared for later steps. It takes about 2 mins for each pileup job.
$GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101035.recal.bam > 121101035.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101043.recal.bam > 121101043.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101050.recal.bam > 121101050.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101052.recal.bam > 121101052.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101415.recal.bam > 121101415.recal.pileup & # $GC/bin/samtools mpileup -q 30 -Q 20 -f $REF/human.g1k.v37.fa -l $HGDP/HGDP_938.bed $BAM/121101861.recal.bam > 121101861.recal.pileup &
We use -q 30 and -Q 20 to exclude reads that have mapping quality score lower than 30 or base quality score lower than 20.
Step 2: pileup --> seq
In this step, we will generate a file called "hapmap_trios.seq", containing the information of 6 samples. It takes about 30 seconds to run. We will use the pre-generated pileup files in the $BAM folder.
python ./LASER-2.01/pileup2seq/pileup2seq.py \ -m $HGDP/HGDP_938.site \ -b $BAM/AMD_roi_1-based.bed \ -i $BAM/AMD_hapmap_trios_id.txt \ -o hapmap_trios \ $BAM/121101035.recal.pileup \ $BAM/121101043.recal.pileup \ $BAM/121101050.recal.pileup \ $BAM/121101052.recal.pileup \ $BAM/121101415.recal.pileup \ $BAM/121101861.recal.pileup &
In the above command, -b provides the targeted regions to exclude and -i specifies alternative IDs for the BAM files to be used in the .seq file (including popID and indivID). -b and -i are optional.
Estimating ancestry coordinates
Step 0: Generate the reference ancestry space
LASER can perform principal components analysis (PCA) on genotype data of the reference panel to generate a reference ancestry space.
# ./LASER-2.01/laser -g $HGDP/HGDP_938.geno -pca 1 -k 30 -o HGDP_938
The above command takes ~20 minutes to finish. We will skip this step, and use a set of reference ancestry coordinates that have been generated in the file $HGDP/HGDP_938.RefPC.coord. View the reference coordinates:
less -S $HGDP/HGDP_938.RefPC.coord
Step 1: Estimate ancestry for sequenced samples
Submit two jobs to place sequenced samples into the reference ancestry space:
./LASER-2.01/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s hapmap_trios.seq -K 20 -k 4 -x 1 -y 3 -o hapmap_trios.1-3 & ./LASER-2.01/laser -g $HGDP/HGDP_938.geno -c $HGDP/HGDP_938.RefPC.coord -s hapmap_trios.seq -K 20 -k 4 -x 4 -y 6 -o hapmap_trios.4-6 &
The first job will process samples 1 to 3 and the second job will processed samples 4 to 6. Each sequenced sample will be projected from a 20-dimensional PCA space onto a 4-dimensional reference ancestry space. The running time is ~10 minutes for processing 3 samples in each job.
Step 2: Combine results
Results from previous step will be output to two files "hapmap_trios.1-3.SeqPC.coord" and "hapmap_trios.4-6.SeqPC.coord". Here we simply concatenate the two files while skipping the header line of the second file.
cp hapmap_trios.1-3.SeqPC.coord hapmap_trios.SeqPC.coord more +2 hapmap_trios.4-6.SeqPC.coord >> hapmap_trios.SeqPC.coord
View the results:
less -S hapmap_trios.SeqPC.coord
The results should look like below:
popID indivID L1 Ci K t PC1 PC2 PC3 PC4 YRI NA19238 78386 0.170864 20 0.999688 467.989 -210.294 -14.1729 -14.4204 CEU NA12892 85486 0.185973 20 0.999723 10.796 199.095 -9.90387 -21.4534 CEU NA12891 87588 0.190442 20 0.99973 2.04224 196.07 -19.5705 -12.8022 CEU NA12878 83213 0.181748 20 0.999711 4.34591 199.861 -12.4825 -22.6281 YRI NA19239 87564 0.193424 20 0.999734 474.464 -215.96 -9.02921 -19.7372 YRI NA19240 95866 0.213874 20 0.999748 469.914 -214.94 -14.9923 -13.6559
Visualizing results
Example R codes are available in ./LASER-2.01/plot/. Let's copy the folder to current working directory:
cp -r ./LASER-2.01/plot/ ./
Go to the plot folder and run the script to plot results:
cd plot Rscript plotHGDP.r $HGDP/HGDP_938.RefPC.coord ../hapmap_trios.SeqPC.coord
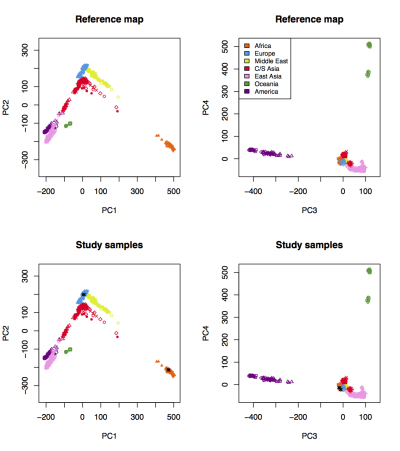
A figure named "Results_on_HGDP.pdf" will be generated. Visualize the figure:
evince Results_on_HGDP.pdf &
We expect to see the following figure, in which 3 CEU samples cluster with HGDP Europeans and 3 YRI samples cluster with HGDP Africans: