Difference between revisions of "SeqShop: Genetic Association Analysis Practical, December 2014"
| (18 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
== Introduction == | == Introduction == | ||
| + | Main Workshop wiki page: [[SeqShop: May 2015]] | ||
| + | |||
[[Media:Seqshop association 2014 06.pdf|View Lecture Slides]] | [[Media:Seqshop association 2014 06.pdf|View Lecture Slides]] | ||
| Line 13: | Line 15: | ||
== Setup in person at the SeqShop Workshop == | == Setup in person at the SeqShop Workshop == | ||
''This section is specifically for the SeqShop Workshop computers.'' | ''This section is specifically for the SeqShop Workshop computers.'' | ||
| − | <div class="mw-collapsible" style="width:600px"> | + | <div class="mw-collapsible mw-collapsed" style="width:600px"> |
''If you are not running during the SeqShop Workshop, please skip this section.'' | ''If you are not running during the SeqShop Workshop, please skip this section.'' | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
| + | <div class="mw-collapsible mw-collapsed" style="width:600px"> | ||
| + | ''If you are not already logged in, please expand this section.'' | ||
| + | <div class="mw-collapsible-content"> | ||
{{SeqShopLogin}} | {{SeqShopLogin}} | ||
| + | </div> | ||
| + | </div> | ||
=== Setup your run environment=== | === Setup your run environment=== | ||
| Line 27: | Line 34: | ||
* Setup an output directory | * Setup an output directory | ||
** It will leave your output directory from the previous tutorial in tact. | ** It will leave your output directory from the previous tutorial in tact. | ||
| − | source /home/ | + | source /net/seqshop-server/home/mktrost/seqshop/setup.txt |
* You won't see any output after running <code>source</code> | * You won't see any output after running <code>source</code> | ||
** It silently sets up your environment | ** It silently sets up your environment | ||
** If you want to view the detail of the setup, type | ** If you want to view the detail of the setup, type | ||
| − | less /home/mktrost/seqshop/setup.txt | + | less /net/seqshop-server/home/mktrost/seqshop/setup.txt |
and press 'q' to finish. | and press 'q' to finish. | ||
| Line 37: | Line 44: | ||
View setup.txt | View setup.txt | ||
<div class="mw-collapsible-content" style="width:800px"> | <div class="mw-collapsible-content" style="width:800px"> | ||
| − | export GC=/ | + | export GC=/net/seqshop-server/home/mktrost/seqshop/gotcloud |
| − | + | export SS=/net/seqshop-server/home/mktrost/seqshop/example | |
| − | export | + | export EPACTS=/net/seqshop-server/home/mktrost/seqshop/epacts |
| − | |||
| − | export | ||
| − | |||
| − | |||
export OUT=~/out | export OUT=~/out | ||
mkdir -p ${OUT} | mkdir -p ${OUT} | ||
| Line 50: | Line 53: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
== Setup when running on your own outside of the SeqShop Workshop == | == Setup when running on your own outside of the SeqShop Workshop == | ||
''This section is specifically for running on your own outside of the SeqShop Workshop.'' | ''This section is specifically for running on your own outside of the SeqShop Workshop.'' | ||
| − | <div class="mw-collapsible | + | <div class="mw-collapsible" style="width:600px"> |
''If you are running during the SeqShop Workshop, please skip this section.'' | ''If you are running during the SeqShop Workshop, please skip this section.'' | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
| Line 66: | Line 68: | ||
cd ~/seqshop/ | cd ~/seqshop/ | ||
* download, decompress, and build the version of epacts that was tested with this tutorial: | * download, decompress, and build the version of epacts that was tested with this tutorial: | ||
| − | wget http:// | + | wget http://csg.sph.umich.edu//kang/epacts/download/EPACTS-3.2.6.tar.gz |
tar xvf EPACTS-3.2.6.tar.gz | tar xvf EPACTS-3.2.6.tar.gz | ||
cd EPACTS-3.2.6 | cd EPACTS-3.2.6 | ||
| Line 247: | Line 249: | ||
Let's run a single-variant association analysis using a score test. | Let's run a single-variant association analysis using a score test. | ||
| − | $EPACTS/bin/epacts | + | $EPACTS/bin/epacts single --ped $SS/assoc/seqshop.ped --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/single --region 22:36000000-37000000 --test b.score --min-mac 1 --pheno PHENO --run 2 |
After running it, you will see EPACTS output files by looking at | After running it, you will see EPACTS output files by looking at | ||
| Line 267: | Line 269: | ||
22 36987861 36987861 22:36987861_A/G 62 31 1 0.25 2.0898e-06 4.7445 31 31 0.43548 0.064516 | 22 36987861 36987861 22:36987861_A/G 62 31 1 0.25 2.0898e-06 4.7445 31 31 0.43548 0.064516 | ||
22 36985499 36985499 22:36985499_C/T 62 29 1 0.23387 5.7389e-06 4.5358 31 31 0.40323 0.064516 | 22 36985499 36985499 22:36985499_C/T 62 29 1 0.23387 5.7389e-06 4.5358 31 31 0.40323 0.064516 | ||
| + | 22 36998907 36998907 22:36998907_C/T 62 58 1 0.46774 1.3679e-05 -4.3489 31 31 0.25806 0.67742 | ||
22 36978260 36978260 22:36978260_G/T 62 28 1 0.22581 1.5051e-05 4.3279 31 31 0.3871 0.064516 | 22 36978260 36978260 22:36978260_G/T 62 28 1 0.22581 1.5051e-05 4.3279 31 31 0.3871 0.064516 | ||
| − | 22 | + | 22 36667082 36667082 22:36667082_T/G 62 27 1 0.21774 0.00015573 -3.7817 31 31 0.064516 0.37097 |
| − | 22 | + | </div> |
| + | </div> | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" style="width:400px"> | ||
| + | Interpretation for top associated variant chr22:36995620_A/G: | ||
| + | <div class="mw-collapsible-content" style="width:800px"> | ||
| + | #CHROM BEGIN END MARKER_ID NS AC CALLRATE MAF PVALUE SCORE NS.CASE NS.CTRL AF.CASE AF.CTRL | ||
| + | 22 36995620 36995620 22:36995620_A/G 62 36 1 0.29032 5.6717e-09 5.8262 31 31 0.51613 0.064516 | ||
| + | |||
| + | The score test PVALUE = 5.6717e-09 is strongly significant. | ||
| + | For the notation chr22:36995620_A/G, the reference allele is the first allele "A", and the non-reference (effect) allele is the second allele "G". | ||
| + | The SCORE statistic = 0.29032 is positive (e.g. SCORE>0), meaning that the effect allele INCREASES your risk for disease. Note that the SCORE is NOT the BETA or log odds ratio. To calculate the BETA and SE(BETA), you must run the b.wald test. | ||
| + | There is a large difference between the allele frequencies for cases and controls. AF.CASE = 0.51613 >> AF.CTRL = 0.0064516. | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 313: | Line 328: | ||
Then the results may look similar to previous ones. | Then the results may look similar to previous ones. | ||
| − | + | head $OUT/assoc/emmax.epacts.top5000 | |
== Run Groupwise Test == | == Run Groupwise Test == | ||
| Line 323: | Line 338: | ||
The group file is simply a list of marker per group name, as shown below. | The group file is simply a list of marker per group name, as shown below. | ||
| − | cat $OUT/assoc/snps.anno.grp | + | cat $OUT/assoc/snps.anno.grp |
| + | |||
APOL1 22:36655735_G/A 22:36657740_G/A 22:36661330_G/A 22:36661566_G/A 22:36661646_G/A 22:36661891_G/A 22:36661906_A/G | APOL1 22:36655735_G/A 22:36657740_G/A 22:36661330_G/A 22:36661566_G/A 22:36661646_G/A 22:36661891_G/A 22:36661906_A/G | ||
APOL2 22:36623731_T/C 22:36623920_G/A 22:36629466_T/A 22:36633107_C/A | APOL2 22:36623731_T/C 22:36623920_G/A 22:36629466_T/A 22:36633107_C/A | ||
| Line 347: | Line 363: | ||
View Output file | View Output file | ||
<div class="mw-collapsible-content" style="width:800px"> | <div class="mw-collapsible-content" style="width:800px"> | ||
| − | #CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE | + | #CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE BETA SEBETA ZSTAT |
| − | 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0. | + | 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0.28783 0.80648 0.75876 1.0629 |
| − | 22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0. | + | 22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0.99286 -17.704 1978.1 -0.0089502 |
| − | 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0. | + | 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0.643 -0.44056 0.95048 -0.46351 |
| − | 22 36587154 36598081 22:36587154-36598081_APOL4 62 0. | + | 22 36587154 36598081 22:36587154-36598081_APOL4 62 0.096774 12 4 0 0.3989 0.76461 0.90638 0.84358 |
| − | 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0. | + | 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0.076266 1.9741 1.1136 1.7728 |
| − | 22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA | + | 22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA NA NA |
| − | 22 36681163 36710183 22:36681163-36710183_MYH9 62 0. | + | 22 36681163 36710183 22:36681163-36710183_MYH9 62 0.048387 3 1 0 0.9904 16.668 1385.4 0.012031 |
| − | 22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA | + | 22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA NA NA |
| − | 22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 | + | 22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 5.9095e-16 1.4376 4.1107e-16 |
| − | 22 36792162 36792162 22:36792162-36792162_RP4-633O19__A.1 NA NA 1 0 0 NA NA | + | 22 36792162 36792162 22:36792162-36792162_RP4-633O19__A.1 NA NA 1 0 0 NA NA NA NA |
</div> | </div> | ||
</div> | </div> | ||
| Line 373: | Line 389: | ||
<div class="mw-collapsible-content" style="width:800px"> | <div class="mw-collapsible-content" style="width:800px"> | ||
#CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE STATRHO | #CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE STATRHO | ||
| − | 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0. | + | 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0.22427 1 |
22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0.038657 NA | 22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0.038657 NA | ||
| − | 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0. | + | 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0.40763 0 |
| − | 22 36587154 36598081 22:36587154-36598081_APOL4 62 0. | + | 22 36587154 36598081 22:36587154-36598081_APOL4 62 0.096774 12 4 0 0.55686 1 |
| − | 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0. | + | 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0.15235 0.3 |
22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA | 22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA | ||
| − | 22 36681163 36710183 22:36681163-36710183_MYH9 62 0. | + | 22 36681163 36710183 22:36681163-36710183_MYH9 62 0.048387 3 1 0 0.075809 NA |
22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA | 22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA | ||
22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 NA | 22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 NA | ||
| Line 385: | Line 401: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | == Return to Workshop Wiki Page == | ||
| + | Return to main workshop wiki page: [[SeqShop: December 2014]] | ||
Latest revision as of 10:27, 2 February 2017
Introduction
Main Workshop wiki page: SeqShop: May 2015
View Introductory Slides for Practical Session
Goals of This Session
- Understand how to annotate variants using EPACTS
- Understand how to run single variant association analysis using EPACTS
- Understand how to run rare variant association test using EPACTS
- Understand how to visualize the association output from EPACTS
Setup in person at the SeqShop Workshop
This section is specifically for the SeqShop Workshop computers.
If you are not running during the SeqShop Workshop, please skip this section.
If you are not already logged in, please expand this section.
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
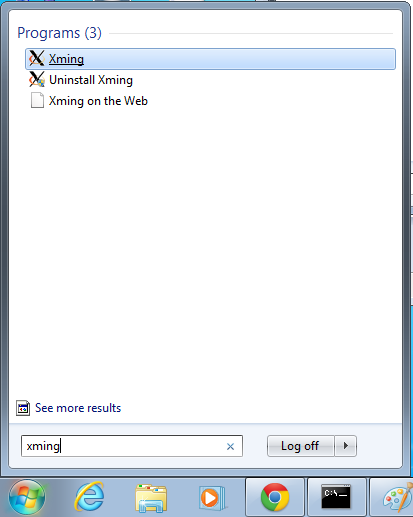
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
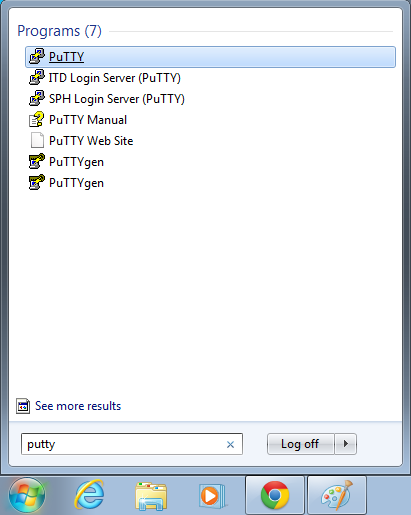
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
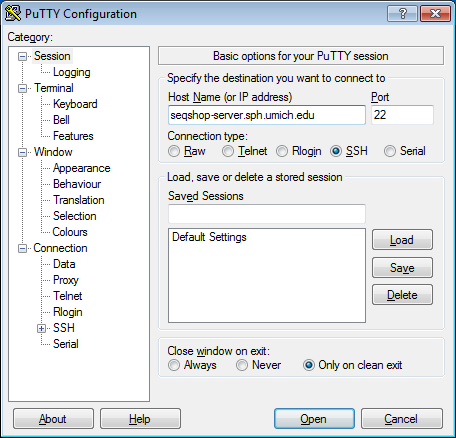
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
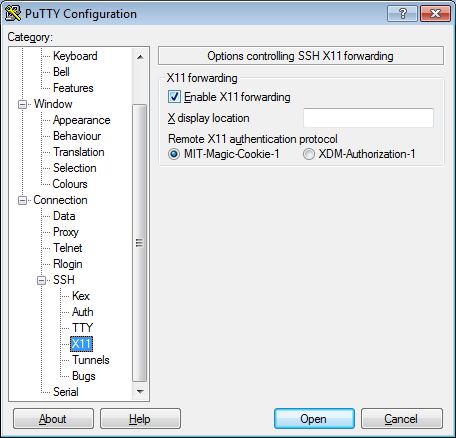
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup your run environment
This is the same setup you did for the previous tutorial, but you need to redo it each time you log in.
This will setup some environment variables to point you to
- GotCloud program
- Tutorial input files
- Setup an output directory
- It will leave your output directory from the previous tutorial in tact.
source /net/seqshop-server/home/mktrost/seqshop/setup.txt
- You won't see any output after running
source- It silently sets up your environment
- If you want to view the detail of the setup, type
less /net/seqshop-server/home/mktrost/seqshop/setup.txt
and press 'q' to finish.
View setup.txt
export GC=/net/seqshop-server/home/mktrost/seqshop/gotcloud
export SS=/net/seqshop-server/home/mktrost/seqshop/example
export EPACTS=/net/seqshop-server/home/mktrost/seqshop/epacts
export OUT=~/out
mkdir -p ${OUT}
Setup when running on your own outside of the SeqShop Workshop
This section is specifically for running on your own outside of the SeqShop Workshop.
If you are running during the SeqShop Workshop, please skip this section.
This tutorial builds on the alignment & snpcall tutorials, if you have not already, please first run those tutorials: Alignment Tutorial & SNP Calling Tutorial
Download & Build EPACTS
If you do not already have EPACTS:
- cd to where you want EPACTS installed (you can change this to any directory you want)
mkdir -p ~/seqshop cd ~/seqshop/
- download, decompress, and build the version of epacts that was tested with this tutorial:
wget http://csg.sph.umich.edu//kang/epacts/download/EPACTS-3.2.6.tar.gz tar xvf EPACTS-3.2.6.tar.gz cd EPACTS-3.2.6 ./configure --prefix=$HOME/seqshop/epacts make make install cd ../..
Setup your run environment
Environment variables will be used throughout the tutorial.
We recommend that you setup these variables so you won't have to modify every command in the tutorial.
- Point to where you installed GotCloud
- Point to where you installed the seqshop files
- Point to where you want the output to go
- Using bash (replace the paths below with the appropriate paths):
export GC=~/seqshop/gotcloud export SS=~/seqshop/example export OUT=~/seqshop/output
- Using tcsh (replace the paths below with the appropriate paths):
setenv GC ~/seqshop/gotcloud setenv SS ~/seqshop/example setenv OUT ~/seqshop/output
- Additional variables for EPACTS:
- Using bash (replace the paths below with the appropriate paths):
export EPACTS=~/seqshop/epacts
- Using tcsh (replace the paths below with the appropriate paths):
setenv EPACTS ~/seqshop/epacts
Preparing Input Files
Input VCF file
We will use SNP genotypes from the SNP calling session, after LD-aware genotype refinement. Check the contents of the VCF file using the following command.
zless ${OUT}/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz
Phenotype Information
Phenotype information is prepared in PED format commonly used in other GWAS software such as MERLIN or PLINK.
less ${SS}/assoc/seqshop.ped
The first several line should look like below.
View Data
#FAM_ID IND_ID DAD_ID MOM_ID SEX PHENO HG00551 HG00551 0 0 0 0 HG00553 HG00553 0 0 0 0 HG00554 HG00554 0 0 0 0 HG00637 HG00637 0 0 0 0 HG00638 HG00638 0 0 0 0 HG00640 HG00640 0 0 0 1 HG00641 HG00641 0 0 0 1 HG00734 HG00734 0 0 0 1 HG00736 HG00736 0 0 0 0 ...
Binary phenotype can be encoded as 0-1 or 1-2. If the column contains more than two distinct values, it will automatically be recognized as quantitative values.
EPACTS allows PED file to have a header line. The header line should contain the description of each column. EPACTS also accepts a standard PED format where .ped file contains the phenotype data and .dat file contains the information about each column.
Installed version of EPACTS
EPACTS are installed in the server. If you want to install EPACTS by yourself, visit EPACTS page for more details
ls $EPACTS/bin
View EPACTS executable files
anno epacts epacts-cis-extract epacts-group epacts-multi epacts.pm epstopdf test_run_epacts.sh bgzip epacts-anno epacts-download epacts-make-group epacts-pca-plot epacts-single pEmmax vcfast chaps epacts-cat epacts-enrich epacts-make-kin epacts-plot epacts-zoom tabix wGetOptions.pm
Note that some tools undocumented in EPACTS documentation is under development and may not work.
Annotating Variants with EPACTS
There are multiple software tools that provides a function to annotate variants, such as Variant Effect Predictor (VEP) that is used in 1000 Genomes Project. While most annotation software provides very similar results to each other, their computational efficiency can substantially vary. The annotation software EPACTS provides is extremely fast and can provide genome-wide annotation results in orders of magnitude faster than other widely available annotation software.
In order to annotate variants with EPACTS, one can use epacts-anno module.
mkdir --p $OUT/assoc $EPACTS/bin/epacts-anno --in $OUT/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz --out $OUT/assoc/snps.anno.vcf.gz --ref $SS/ref22/human.g1k.v37.chr22.fa
Then you will see a series of messages before annotation finishes.
View the expected messages
/home/hmkang/seqshop/epacts/bin/anno -i /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz -r \
/home/hmkang/seqshop/ref22/human_g1k_v37.chr22.fa -f refGene -g /home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz \
-c /home/hmkang/seqshop/epacts/share/EPACTS/codon.txt -o /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz --inputFormat vcf \
-p /home/hmkang/seqshop/epacts/share/EPACTS/priority.txt
The following parameters are available. Ones with "[]" are in effect:
Available Options
Required Parameters :
-i [/net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz]
-o [/net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz]
Gene Annotation
Parameters : -g [/home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz]
-r [/home/hmkang/seqshop/ref22/human_g1k_v37.chr22.fa]
--inputFormat [vcf], --checkReference, -f [refGene]
-p [/home/hmkang/seqshop/epacts/share/EPACTS/priority.txt]
-c [/home/hmkang/seqshop/epacts/share/EPACTS/codon.txt]
-u [], -d [], --se [], --si [], --outputFormat []
Other Annotation Tools : --genomeScore [], --bed [], --tabix []
Load reference genome /home/hmkang/seqshop/ref22/human_g1k_v37.chr22.fa...
DONE: 1 chromosomes and 51304566 bases are loaded.
Load codon file /home/hmkang/seqshop/epacts/share/EPACTS/codon.txt...
DONE: codon file loaded.
Load priority file /home/hmkang/seqshop/epacts/share/EPACTS/priority.txt...
DONE: 24 priority annotation types loaded.
Load gene file /home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz...
DONE: 92627 gene loaded.
DONE: Generated frequency of each annotype type in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.anno.frq ].
DONE: Generated frequency of each highest priority annotation type in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.top.anno.frq ].
Ts/Tv ratio: 2.35733
Ts observed: 2718 times; Tv observed: 1153 times.
DONE: Generated frequency of each base change in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.base.frq ].
DONE: Generated frequency of each codon change in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.codon.frq ].
DONE: Generated frequency of indel length in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.indel.frq ].
..............................................
... Anno(tation) ...
... Xiaowei Zhan, Goncalo Abecasis ...
... Speical Thanks: ...
... Hyun Ming Kang, Yanming Li ...
... zhanxw@umich.edu ...
... Sep 2011 ...
................................................
DONE: 3871 varaints are annotated.
DONE: Generated annotation output in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz ].
Annotation succeed!
mv /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp
/home/hmkang/seqshop/epacts/bin/bgzip -c /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp > /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz
/home/hmkang/seqshop/epacts/bin/tabix -pvcf -f /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz
rm /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp
rm /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.log /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.top.anno.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.anno.frq /net/seqshop- server/hmkang/out/assoc/snps.anno.vcf.gz.base.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.codon.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.indel.frq
After running annotation, you can check the annotation results. Let's look at the APOL g1 risk allele we manually examined in the SNP calling section.
$GC/bin/tabix $OUT/assoc/snps.anno.vcf.gz 22:36661906 | head -1 | cut -f 1-8
View the annotation results
22 36661906 . A G 18 PASS DP=409;MQ=59;NS=62;AN=124;AC=2;AF=0.013827;AB=0.4065;AZ=-0.5287;FIC=-0.0092;
SLRT=-0.0075;HWEAF=0.0138;HWDAF=0.0276,0.0000;LBS=36,36,0,0,1,1,0,0;OBS=145,191,0,0,3,2,0,0;STR=-0.040;
STZ=-0.740;CBR=0.008;CBZ=0.144;IOR=0.000;IOZ=-1.370;AOI=-5.614;AOZ=-4.243;LQR=0.178;MQ0=0.000;MQ10=0.000;MQ20=0.000;
MQ30=0.000;SVM=1.51214;BAVGPOST=0.998;BRSQ=0.941;LDAF=0.0161;AVGPOST=1.0000;RSQ=1.0000;ERATE=0.0019;THETA=0.0013;
ANNO=Nonsynonymous:APOL1;ANNOFULL=APOL1/ENST00000397278.3:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/6):Exon|
APOL1/ENST00000426053.1:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base971/1143:Codon324/381:Exon5/5):Exon|
APOL1/ENST00000422706.1:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/6):Exon|
APOL1/ENST00000319136.4:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1073/1245:Codon358/415:Exon7/7):Exon|
APOL1/ENST00000347595.7:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base662/834:Codon221/278:Exon3/3):Exon|
APOL1/ENST00000397279.4:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/7):Exon
- What is the function of this variant?
- How many different transcript does the variant overlap with?
- How can you represent the variant in terms of amino acid changes?
Single Variant Association Analysis
Let's run a single-variant association analysis using a score test.
$EPACTS/bin/epacts single --ped $SS/assoc/seqshop.ped --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/single --region 22:36000000-37000000 --test b.score --min-mac 1 --pheno PHENO --run 2
After running it, you will see EPACTS output files by looking at
ls $OUT/assoc
The top association results can be viewed by
head $OUT/assoc/single.epacts.top5000
View top association results
#CHROM BEGIN END MARKER_ID NS AC CALLRATE MAF PVALUE SCORE NS.CASE NS.CTRL AF.CASE AF.CTRL 22 36995620 36995620 22:36995620_A/G 62 36 1 0.29032 5.6717e-09 5.8262 31 31 0.51613 0.064516 22 36993088 36993088 22:36993088_G/C 62 30 1 0.24194 7.3258e-07 4.9525 31 31 0.43548 0.048387 22 36997871 36997871 22:36997871_G/T 62 30 1 0.24194 7.3258e-07 4.9525 31 31 0.43548 0.048387 22 36987368 36987368 22:36987368_G/A 62 31 1 0.25 2.0898e-06 4.7445 31 31 0.43548 0.064516 22 36987861 36987861 22:36987861_A/G 62 31 1 0.25 2.0898e-06 4.7445 31 31 0.43548 0.064516 22 36985499 36985499 22:36985499_C/T 62 29 1 0.23387 5.7389e-06 4.5358 31 31 0.40323 0.064516 22 36998907 36998907 22:36998907_C/T 62 58 1 0.46774 1.3679e-05 -4.3489 31 31 0.25806 0.67742 22 36978260 36978260 22:36978260_G/T 62 28 1 0.22581 1.5051e-05 4.3279 31 31 0.3871 0.064516 22 36667082 36667082 22:36667082_T/G 62 27 1 0.21774 0.00015573 -3.7817 31 31 0.064516 0.37097
Interpretation for top associated variant chr22:36995620_A/G:
#CHROM BEGIN END MARKER_ID NS AC CALLRATE MAF PVALUE SCORE NS.CASE NS.CTRL AF.CASE AF.CTRL 22 36995620 36995620 22:36995620_A/G 62 36 1 0.29032 5.6717e-09 5.8262 31 31 0.51613 0.064516
The score test PVALUE = 5.6717e-09 is strongly significant. For the notation chr22:36995620_A/G, the reference allele is the first allele "A", and the non-reference (effect) allele is the second allele "G". The SCORE statistic = 0.29032 is positive (e.g. SCORE>0), meaning that the effect allele INCREASES your risk for disease. Note that the SCORE is NOT the BETA or log odds ratio. To calculate the BETA and SE(BETA), you must run the b.wald test. There is a large difference between the allele frequencies for cases and controls. AF.CASE = 0.51613 >> AF.CTRL = 0.0064516.
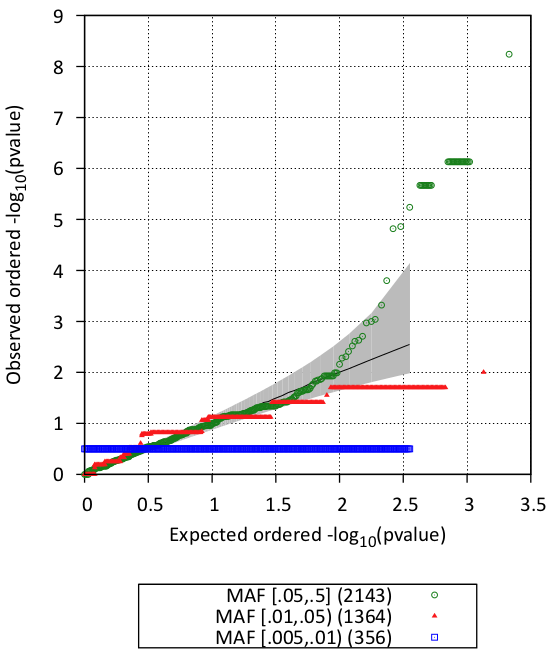
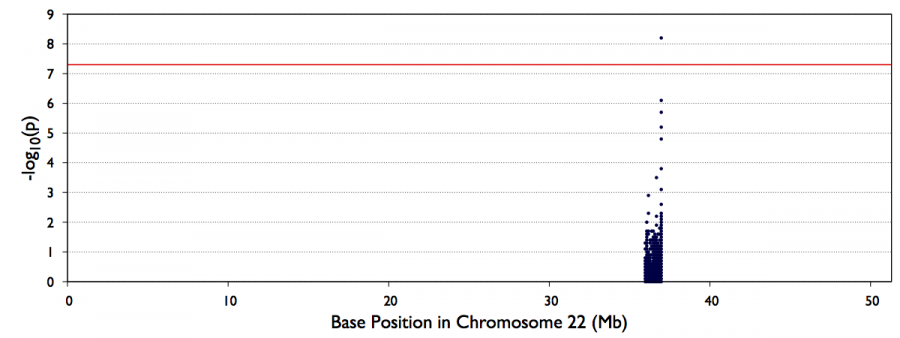
You can look also visualize the results by QQ-plot and Manhattan plot
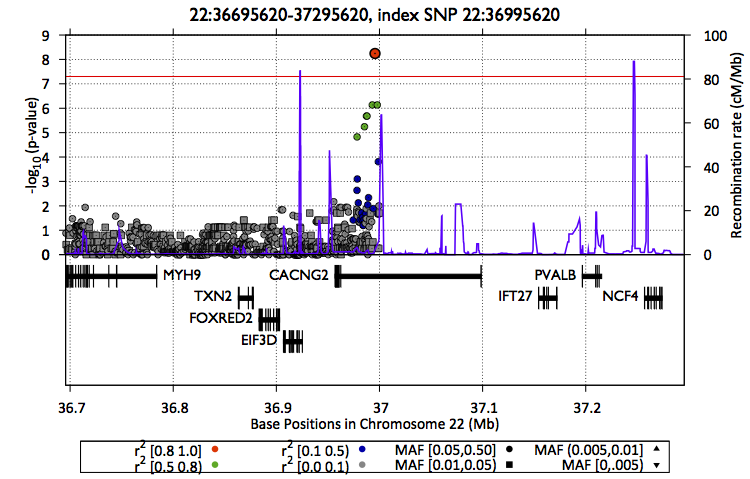
Also, you can create a zoom plot focusing on the region of interest
$EPACTS/bin/epacts-zoom --vcf $OUT/assoc/snps.anno.vcf.gz --pos 22:36995620 --prefix $OUT/assoc/single
If you want to run EMMAX, you first need to create a kinship matrix
$EPACTS/bin/epacts-make-kin --vcf $OUT/assoc/snps.anno.vcf.gz --min-maf 0.01 --out $OUT/assoc/snps.anno.kinf --run 2 --chr 22
And run EMMAX test specifying the kinship matrix
$EPACTS/bin/epacts-single --ped $SS/assoc/seqshop.ped --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/emmax --region 22:36000000-37000000 --test q.emmax --pheno PHENO --run 2 --kinf $OUT/assoc/snps.anno.kinf
Then the results may look similar to previous ones.
head $OUT/assoc/emmax.epacts.top5000
Run Groupwise Test
To run group-wise test such as gene-level burden test, you need to create a marker group file. If your VCF is already annotated, you can create a group file by running
$EPACTS/bin/epacts make-group --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/snps.anno.grp --nonsyn
The group file is simply a list of marker per group name, as shown below.
cat $OUT/assoc/snps.anno.grp
APOL1 22:36655735_G/A 22:36657740_G/A 22:36661330_G/A 22:36661566_G/A 22:36661646_G/A 22:36661891_G/A 22:36661906_A/G APOL2 22:36623731_T/C 22:36623920_G/A 22:36629466_T/A 22:36633107_C/A APOL3 22:36537763_C/T 22:36537798_G/A 22:36556768_G/A 22:36556823_G/T APOL4 22:36587154_G/T 22:36587202_G/A 22:36587223_G/T 22:36587346_C/T 22:36587511_C/T 22:36587704_T/C 22:36587886_C/T 22:36593714_G/A 22:36597744_A/C 22:36598049_C/G 22:36598058_T/C 22:36598081_A/T APOL5 22:36122356_G/A 22:36122380_T/A 22:36122930_C/T 22:36123083_C/T 22:36124860_C/G FOXRED2 22:36900271_T/C 22:36900806_A/G MYH9 22:36681163_G/C 22:36684354_T/C 22:36710183_T/C Metazoa_SRP 22:36711990_C/G RBFOX2 22:36424450_A/C RP4-633O19__A.1 22:36792162_G/A
If you have your own annotation, you can create your own burden test unit by modifying this file.
If you want to run a collapsing burden test (CMC), run the following command
$EPACTS/bin/epacts group --ped $SS/assoc/seqshop.ped --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/group.collapse --test b.collapse --groupf $OUT/assoc/snps.anno.grp --pheno PHENO --run 2
You can view the results by examining the output file
cat $OUT/assoc/group.collapse.epacts
View Output file
#CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE BETA SEBETA ZSTAT 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0.28783 0.80648 0.75876 1.0629 22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0.99286 -17.704 1978.1 -0.0089502 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0.643 -0.44056 0.95048 -0.46351 22 36587154 36598081 22:36587154-36598081_APOL4 62 0.096774 12 4 0 0.3989 0.76461 0.90638 0.84358 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0.076266 1.9741 1.1136 1.7728 22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA NA NA 22 36681163 36710183 22:36681163-36710183_MYH9 62 0.048387 3 1 0 0.9904 16.668 1385.4 0.012031 22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA NA NA 22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 5.9095e-16 1.4376 4.1107e-16 22 36792162 36792162 22:36792162-36792162_RP4-633O19__A.1 NA NA 1 0 0 NA NA NA NA
You can run SKAT-O test in a similar way, but with a special tag
$EPACTS/bin/epacts group --ped $SS/assoc/seqshop.ped --vcf $OUT/assoc/snps.anno.vcf.gz --out $OUT/assoc/group.skato --test skat --skat-o --groupf $OUT/assoc/snps.anno.grp --pheno PHENO --run 2
And view output files
cat $OUT/assoc/group.skato.epacts
View Output file
#CHROM BEGIN END MARKER_ID NS FRAC_WITH_RARE NUM_ALL_VARS NUM_PASS_VARS NUM_SING_VARS PVALUE STATRHO 22 36655735 36661906 22:36655735-36661906_APOL1 62 0.14516 7 4 0 0.22427 1 22 36623731 36633107 22:36623731-36633107_APOL2 62 0.064516 4 1 0 0.038657 NA 22 36537763 36556823 22:36537763-36556823_APOL3 62 0.080645 4 2 0 0.40763 0 22 36587154 36598081 22:36587154-36598081_APOL4 62 0.096774 12 4 0 0.55686 1 22 36122356 36124860 22:36122356-36124860_APOL5 62 0.1129 5 2 0 0.15235 0.3 22 36900271 36900806 22:36900271-36900806_FOXRED2 NA NA 2 0 0 NA NA 22 36681163 36710183 22:36681163-36710183_MYH9 62 0.048387 3 1 0 0.075809 NA 22 36711990 36711990 22:36711990-36711990_Metazoa_SRP NA NA 1 0 0 NA NA 22 36424450 36424450 22:36424450-36424450_RBFOX2 62 0.032258 1 1 0 1 NA 22 36792162 36792162 22:36792162-36792162_RP4-633O19__A.1 NA NA 1 0 0 NA NA
Return to Workshop Wiki Page
Return to main workshop wiki page: SeqShop: December 2014