SeqShop: Genetic Association Analysis Practical, June 2014
(WARNING: Under Construction)
Goals of This Session
- Understand how to annotate variants using EPACTS
- Understand how to run single variant association analysis using EPACTS
- Understand how to run rare variant association test using EPACTS
- Understand how to visualize the association output from EPACTS
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
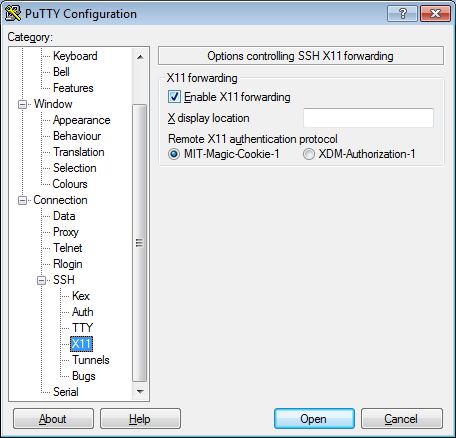
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup your run environment
This is a setup similar to the Structural Variant session.
- GotCloud program
- Tutorial input files
- Setup an output directory
- It will leave your output directory from the previous tutorial in tact.
source /home/hmkang/seqshop/setup.txt
- You won't see any output after running
source- It silently sets up your environment
- If you want to view the detail of the set up, type
less /home/hmkang/seqshop/setup.txt
and press 'q' to finish.
View setup.txt
export GC=/home/hmkang/seqshop/gotcloud
export IN=/home/hmkang/seqshop/inputs
export REF=/home/hmkang/seqshop/reference/chr22
export VTREF=/home/hmkang/seqshop/reference/vtRef
export SV=/home/hmkang/seqshop/reference/svtoolkit
export EXT=/home/hmkang/seqshop/external
export OUT=~/out
mkdir -p ${OUT}
Preparing Input Files
Input VCF file
We will use SNP genotypes from the SNP calling session, after LD-aware genotype refinement. Check the contents of the VCF file using the following command.
zless $OUT/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz
Phenotype Information
Phenotype information is prepared in PED format commonly used in other GWAS software such as MERLIN or PLINK.
less $IN/assoc/seqshop.ped
The first several line should look like below.
View Data
#FAM_ID IND_ID DAD_ID MOM_ID SEX GENO PHENO HG00551 HG00551 0 0 0 1 0 HG00553 HG00553 0 0 0 0 0 HG00554 HG00554 0 0 0 0 0 HG00637 HG00637 0 0 0 0 0 HG00638 HG00638 0 0 0 0 0 HG00640 HG00640 0 0 0 1 1 HG00641 HG00641 0 0 0 2 1 HG00734 HG00734 0 0 0 1 1 HG00736 HG00736 0 0 0 0 0 HG00737 HG00737 0 0 0 0 0 ...
Binary phenotype can be encoded as 0-1 or 1-2. If the column contains more than two distinct values, it will automatically be recognized as quantitative values.
EPACTS allows PED file to have a header line. The header line should contain the description of each column. EPACTS also accepts a standard PED format where .ped file contains the phenotype data and .dat file contains the information about each column.
Installed version of EPACTS
EPACTS are installed in the server. If you want to install EPACTS by yourself, visit EPACTS page for more details
ls $EPACTS/bin
View EPACTS executable files
anno epacts epacts-cis-extract epacts-group epacts-multi epacts.pm epstopdf test_run_epacts.sh bgzip epacts-anno epacts-download epacts-make-group epacts-pca-plot epacts-single pEmmax vcfast chaps epacts-cat epacts-enrich epacts-make-kin epacts-plot epacts-zoom tabix wGetOptions.pm
Note that some tools undocumented in EPACTS documentation is under development and may not work.
Annotating Variants with EPACTS
There are multiple software tools that provides a function to annotate variants, such as Variant Effect Predictor (VEP) that is used in 1000 Genomes Project. While most annotation software provides very similar results to each other, their computational efficiency can substantially vary. The annotation software EPACTS provides is extremely fast and can provide genome-wide annotation results in orders of magnitude faster than other widely available annotation software.
In order to annotate variants with EPACTS, one can use epacts-anno module.
mkdir --p $OUT/assoc $EPACTS/bin/epacts-anno --in $OUT/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz --out $OUT/assoc/snps.anno.vcf.gz
Then you will see a series of messages before annotation finishes.
View the expected messages
/home/hmkang/seqshop/epacts/bin/anno -i /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz -r \
/home/hmkang/seqshop/epacts/share/EPACTS/human_g1k_v37.fasta -f refGene -g /home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz \
-c /home/hmkang/seqshop/epacts/share/EPACTS/codon.txt -o /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz --inputFormat vcf \
-p /home/hmkang/seqshop/epacts/share/EPACTS/priority.txt
The following parameters are available. Ones with "[]" are in effect:
Available Options
Required Parameters :
-i [/net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz]
-o [/net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz]
Gene Annotation
Parameters : -g [/home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz]
-r [/home/hmkang/seqshop/epacts/share/EPACTS/human_g1k_v37.fasta]
--inputFormat [vcf], --checkReference, -f [refGene]
-p [/home/hmkang/seqshop/epacts/share/EPACTS/priority.txt]
-c [/home/hmkang/seqshop/epacts/share/EPACTS/codon.txt]
-u [], -d [], --se [], --si [], --outputFormat []
Other Annotation Tools : --genomeScore [], --bed [], --tabix []
Load reference genome /home/hmkang/seqshop/epacts/share/EPACTS/human_g1k_v37.fasta...
DONE: 84 chromosomes and 3101804739 bases are loaded.
Load codon file /home/hmkang/seqshop/epacts/share/EPACTS/codon.txt...
DONE: codon file loaded.
Load priority file /home/hmkang/seqshop/epacts/share/EPACTS/priority.txt...
DONE: 24 priority annotation types loaded.
Load gene file /home/hmkang/seqshop/epacts/share/EPACTS/hg19_gencodeV14.txt.gz...
DONE: 92627 gene loaded.
DONE: Generated frequency of each annotype type in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.anno.frq ].
DONE: Generated frequency of each highest priority annotation type in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.top.anno.frq ].
Ts/Tv ratio: 2.35733
Ts observed: 2718 times; Tv observed: 1153 times.
DONE: Generated frequency of each base change in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.base.frq ].
DONE: Generated frequency of each codon change in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.codon.frq ].
DONE: Generated frequency of indel length in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.indel.frq ].
..............................................
... Anno(tation) ...
... Xiaowei Zhan, Goncalo Abecasis ...
... Speical Thanks: ...
... Hyun Ming Kang, Yanming Li ...
... zhanxw@umich.edu ...
... Sep 2011 ...
................................................
DONE: 3871 varaints are annotated.
DONE: Generated annotation output in [ /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz ].
Annotation succeed!
mv /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp
/home/hmkang/seqshop/epacts/bin/bgzip -c /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp > /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz
/home/hmkang/seqshop/epacts/bin/tabix -pvcf -f /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz
rm /net/seqshop-server/hmkang/out/thunder/chr22/ALL/thunder/chr22.filtered.PASS.beagled.ALL.thunder.vcf.gz.tmp
rm /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.log /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.top.anno.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.anno.frq /net/seqshop- server/hmkang/out/assoc/snps.anno.vcf.gz.base.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.codon.frq /net/seqshop-server/hmkang/out/assoc/snps.anno.vcf.gz.indel.frq
After running annotation, you can check the annotation results. Let's look at the APOL g1 risk allele we manually examined in the SNP calling section.
$GC/bin/tabix $OUT/assoc/snps.anno.vcf.gz 22:36661906 | head -1 | cut -f 1-8
View the annotation results
22 36661906 . A G 18 PASS DP=409;MQ=59;NS=62;AN=124;AC=2;AF=0.013827;AB=0.4065;AZ=-0.5287;FIC=-0.0092;
SLRT=-0.0075;HWEAF=0.0138;HWDAF=0.0276,0.0000;LBS=36,36,0,0,1,1,0,0;OBS=145,191,0,0,3,2,0,0;STR=-0.040;
STZ=-0.740;CBR=0.008;CBZ=0.144;IOR=0.000;IOZ=-1.370;AOI=-5.614;AOZ=-4.243;LQR=0.178;MQ0=0.000;MQ10=0.000;MQ20=0.000;
MQ30=0.000;SVM=1.51214;BAVGPOST=0.998;BRSQ=0.941;LDAF=0.0161;AVGPOST=1.0000;RSQ=1.0000;ERATE=0.0019;THETA=0.0013;
ANNO=Nonsynonymous:APOL1;ANNOFULL=APOL1/ENST00000397278.3:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/6):Exon|
APOL1/ENST00000426053.1:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base971/1143:Codon324/381:Exon5/5):Exon|
APOL1/ENST00000422706.1:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/6):Exon|
APOL1/ENST00000319136.4:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1073/1245:Codon358/415:Exon7/7):Exon|
APOL1/ENST00000347595.7:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base662/834:Codon221/278:Exon3/3):Exon|
APOL1/ENST00000397279.4:+:Nonsynonymous(AGC/Ser/S->GGC/Gly/G:Base1025/1197:Codon342/399:Exon6/7):Exon
- What is the function of this variant?
- How many different transcript does the variant overlap with?
- How can you represent the variant in terms of amino acid changes?
Run Single Variant Analysis
- Run score test
mkdir ~/out/assoc $S5/epacts/bin/epacts single --ped $S5/examples/index/chr7.CFTR.ped --vcf ~/out/snps/beagle/chr7/chr7.filtered.PASS.beagled.vcf.gz --pheno PHENO --out ~/out/assoc/single.b.score --test b.score --anno --ref $S5/examples/chr7Ref/hs37d5.chr7.fa --region 7:117000000-117500000 --run 1
- Look at the association statistics
head ~/out/assoc/single.b.score.epacts.top5000
- Variant annotation with EPACTS
$S5/epacts/bin/epacts anno --in ~/out/snps/beagle/chr7/chr7.filtered.PASS.beagled.vcf.gz --out ~/out/snps/chr7.filtered.PASS.beagled.anno.vcf.gz --ref $S5/examples/chr7Ref/hs37d5.chr7.fa
Visualizing Zoom Plot
- Use ENCODE GM12878 cell type to diplsy variants in the locusZoom-like plot
$S5/epacts/bin/epacts-zoom --vcf ~/out/snps/chr7.filtered.PASS.beagled.anno.vcf.gz --pos 7:117149147 --prefix ~/out/assoc/single.b.score --cellType Gm12878
Copy the output file single.b.score.zoom.7.117149147.pdf using WinSCP or other file transfer software
Run Groupwise Test
- Create a set of markers to test
$S5/epacts/bin/epacts make-group --vcf ~/out/snps/chr7.filtered.PASS.beagled.anno.vcf.gz --out ~/out/snps/chr7.filtered.PASS.beagled.anno.grp --nonsyn
- Run SKAT-O test
$S5/epacts/bin/epacts group --ped $S5/examples/index/chr7.CFTR.ped --vcf ~/out/snps/chr7.filtered.PASS.beagled.anno.vcf.gz --out ~/out/assoc/group.skat.o --groupf ~/out/snps/chr7.filtered.PASS.beagled.anno.grp --test skat --skat-o --run 2
cat ~/out/assoc/group.skat.o.epacts | cut -f 4,6,10,11
- Run Variable Threshold test
$S5/epacts/bin/epacts group --ped $S5/examples/index/chr7.CFTR.ped --vcf ~/out/snps/chr7.filtered.PASS.beagled.anno.vcf.gz --out ~/out/assoc/group.VT --groupf ~/out/snps/chr7.filtered.PASS.beagled.anno.grp --test VT --run 2
cat ~/out/assoc/group.VT.epacts | cut -f 4,6,10,13