Difference between revisions of "SeqShop: Variant Calling and Filtering for INDELs Practical, June 2014"
| Line 28: | Line 28: | ||
== Examining GotCloud Indel Input files == | == Examining GotCloud Indel Input files == | ||
The GotCloud Indel caller takes the same inputs as GotCloud snpcall. | The GotCloud Indel caller takes the same inputs as GotCloud snpcall. | ||
| + | * BAMs->INDELs rather than BAMs->SNPs | ||
If you want a reminder, of what they look like, here is a link to the previous tutorial : [[SeqShop:_Variant_Calling_and_Filtering_for_SNPs_Practical#Examining_GotCloud_SnpCall_Input_files|GotCloud SnpCall Input Files]] | If you want a reminder, of what they look like, here is a link to the previous tutorial : [[SeqShop:_Variant_Calling_and_Filtering_for_SNPs_Practical#Examining_GotCloud_SnpCall_Input_files|GotCloud SnpCall Input Files]] | ||
| Line 33: | Line 34: | ||
== Running GotCloud Indel == | == Running GotCloud Indel == | ||
${GC}/gotcloud indel --conf ${IN}/gotcloud.conf --numjobs 2 --region 22:36000000-37000000 | ${GC}/gotcloud indel --conf ${IN}/gotcloud.conf --numjobs 2 --region 22:36000000-37000000 | ||
| + | * --numjobs tells GotCloud how many jobs to run in parallel | ||
| + | ** Depends on your system | ||
| + | * --region 22:36000000-37000000 | ||
| + | ** The sample files are just a small region of chromosome 22, so to save time, we tell GotCloud to ignore the other regions | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" style="width:500px"> | ||
| + | Curious if it started running properly? Check out this screenshot: | ||
| + | <div class="mw-collapsible-content"> | ||
| + | [[File:indelStart.png|550px]] | ||
| + | </div> | ||
| + | </div> | ||
| + | This should take about 4-5 minutes to run. | ||
| + | * It should end with a line like: <code>Commands finished in 289 secs with no errors reported</code> | ||
| + | |||
| + | If you cancelled GotCloud part way through, just rerun your GotCloud command and it will pick up where it left off. | ||
| + | |||
| + | |||
| + | == Examining GotCloud indel Ouptut == | ||
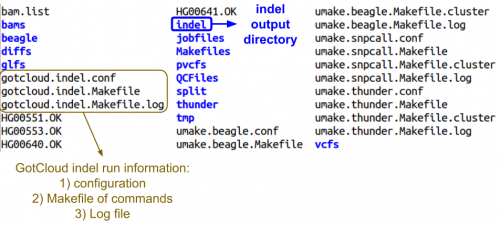
| + | Let's look at the output directory: | ||
| + | ls ${OUT} | ||
| + | |||
| + | ;Do you see any new files or directories? | ||
| + | <div class="mw-collapsible mw-collapsed" style="width:350px"> | ||
| + | * View Annotated Screenshot: | ||
| + | <div class="mw-collapsible-content"> | ||
| + | [[File:indelOut.png|500px]] | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | Let's look at the <code>final</code> directory: | ||
| + | ls ${OUT}/final | ||
| + | Just a <code>chr22</code> directory, so look inside of there: | ||
| + | ls ${OUT}/vcfs/chr22 | ||
| + | ;Can you identify the final indel VCF? | ||
| + | <div class="mw-collapsible mw-collapsed" style="width:350px"> | ||
| + | * Answer & annotated directory listing: | ||
| + | <div class="mw-collapsible-content"> | ||
| + | <ul> | ||
| + | <li>all.genotypes.vcf.gz</li> | ||
| + | </ul> | ||
| + | [[File:indelOutFinal.png|600px]] | ||
| + | </div> | ||
| + | </div> | ||
Revision as of 21:28, 16 June 2014
Goals of This Session
- What we want to learn
- How to generate variant calls for INDELs from BAMs
- How to examine the variants at particular genomic positions
- How to evaluate the quality of INDEL calls
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
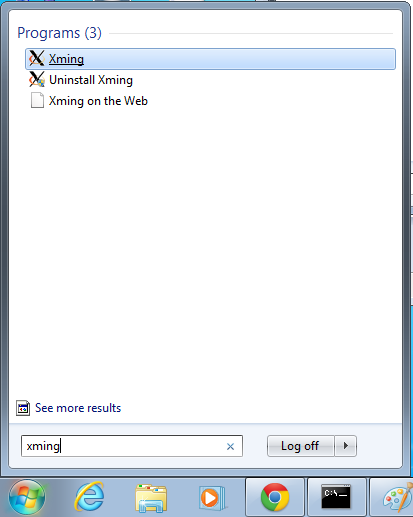
- Start xming so you can open external windows on our Linux machine
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
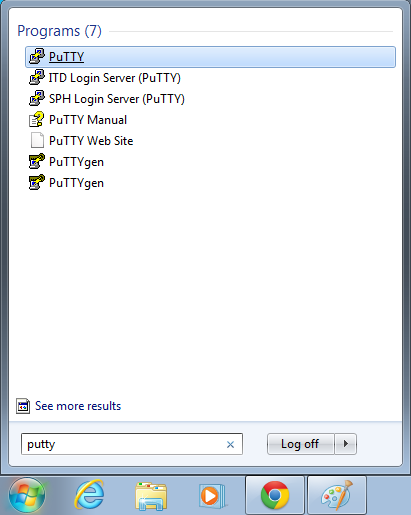
- Open putty
- Start->Enter "putty" in the search and select "PuTTY" from the program list
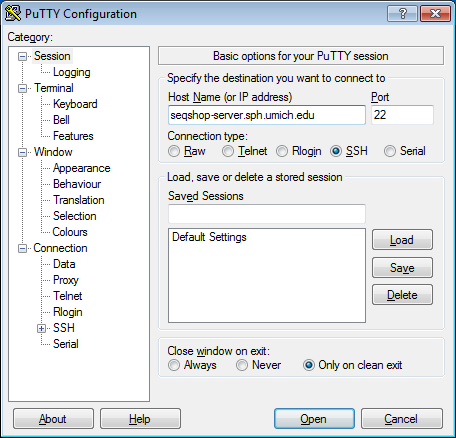
- Configure PuTTY in the PuTTY Configuration window
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
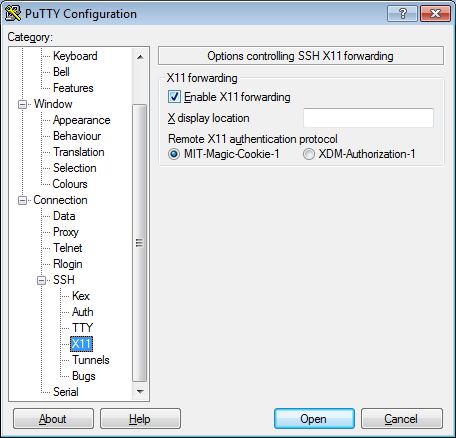
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK - Enter your provided username & password as provided
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup your run environment
This is the same setup you did for the previous tutorial, but you need to redo it each time you log in. This will setup some environment variables to point you to:
- GotCloud program
- Tutorial input files
- Setup an output directory
- It will leave your output directory from the previous tutorial in tact.
source /home/mktrost/seqshop/setup.txt
- You won't see any output after running
source- It silently sets up your environment
Examining GotCloud Indel Input files
The GotCloud Indel caller takes the same inputs as GotCloud snpcall.
- BAMs->INDELs rather than BAMs->SNPs
If you want a reminder, of what they look like, here is a link to the previous tutorial : GotCloud SnpCall Input Files
Running GotCloud Indel
${GC}/gotcloud indel --conf ${IN}/gotcloud.conf --numjobs 2 --region 22:36000000-37000000
- --numjobs tells GotCloud how many jobs to run in parallel
- Depends on your system
- --region 22:36000000-37000000
- The sample files are just a small region of chromosome 22, so to save time, we tell GotCloud to ignore the other regions
This should take about 4-5 minutes to run.
- It should end with a line like:
Commands finished in 289 secs with no errors reported
If you cancelled GotCloud part way through, just rerun your GotCloud command and it will pick up where it left off.
Examining GotCloud indel Ouptut
Let's look at the output directory:
ls ${OUT}
- Do you see any new files or directories?
Let's look at the final directory:
ls ${OUT}/final
Just a chr22 directory, so look inside of there:
ls ${OUT}/vcfs/chr22
- Can you identify the final indel VCF?