Difference between revisions of "Test EPACTS for DIAGRAM"
Clement Ma (talk | contribs) |
|||
| Line 14: | Line 14: | ||
tar xzvf epacts_v2_01.noref_binary.2012_07_06.tar.gz | tar xzvf epacts_v2_01.noref_binary.2012_07_06.tar.gz | ||
| − | = | + | = Accessing help = |
| + | |||
| + | For a list of commands available in EPACTS, type in the following commands: | ||
| + | <pre>'''$ epacts2/epacts help''' | ||
| + | Usage: | ||
| + | epacts [command] [options] | ||
| + | |||
| + | Command: | ||
| + | help Print out brief help message | ||
| + | man Print the full documentation in man page style | ||
| + | single Perform single variant association | ||
| + | group Perform groupwise (burden-style) association test | ||
| + | anno Annotate a VCF file | ||
| + | zoom Create a locus zoom plot from epacts results | ||
| + | meta Perform meta-analysis across multiple epacts results | ||
| + | |||
| + | Visit http://genome.sph.umich.edu/wiki/EPACTS for more detailed documentation | ||
| + | |||
| + | To view options for single variant testing only type in:</pre><pre>$ epacts2/epacts single -help | ||
| + | Usage: | ||
| + | epacts single [options] | ||
| + | |||
| + | Required Options (Run epacts single -man or see wiki for more info): | ||
| + | -vcf STR Input VCF file (tabixed and bgzipped) | ||
| + | -ped STR Input PED file for phenotypes and covariates | ||
| + | -out STR Prefix of output files | ||
| + | -test STR Statistical test to use | ||
| + | |||
| + | ... | ||
| + | |||
| + | |||
| + | </pre> | ||
| + | = Getting started in EPACTS with an example = | ||
Once installed, test out the software by running a quick example using the test data provided in the "example" directory. The example VCF and PED files are: | Once installed, test out the software by running a quick example using the test data provided in the "example" directory. The example VCF and PED files are: | ||
| − | <pre>$ | + | <pre>$ epacts2/example/1000G_exome_chr20_example_softFiltered.calls.vcf.gz |
| − | $ | + | $ epacts2/example/1000G_dummy_pheno.ped |
</pre> | </pre> | ||
<br> Run the single variant score test on the example data using this command: | <br> Run the single variant score test on the example data using this command: | ||
| − | <pre>$ | + | <pre>$ epacts2/epacts single \ |
| − | --vcf | + | --vcf epacts2/example/1000G_exome_chr20_example_softFiltered.calls.vcf.gz \ |
| − | --ped | + | --ped epacts2/example/1000G_dummy_pheno.ped \ |
--min-maf 0.001 --chr 20 --pheno DISEASE --cov AGE --cov SEX --test b.score --anno \ | --min-maf 0.001 --chr 20 --pheno DISEASE --cov AGE --cov SEX --test b.score --anno \ | ||
--out {OUTPUT_DIR}/test --run 2 & | --out {OUTPUT_DIR}/test --run 2 & | ||
| Line 68: | Line 100: | ||
An example Genome-wide manhattan plot (from a genome-wide run) will look like below<br><br> [[Image:Tes b score epacts mh gw.png]] <br> | An example Genome-wide manhattan plot (from a genome-wide run) will look like below<br><br> [[Image:Tes b score epacts mh gw.png]] <br> | ||
| − | + | <pre> | |
| − | |||
| − | |||
| − | |||
| − | <pre> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
...</pre> | ...</pre> | ||
Revision as of 17:12, 26 September 2012
Download EPACTS
EPACTS is available for download here (100Mb) .
Requirements
- Linux 64bit
- Perl 5
Install EPACTS
Uncompress EPACTS package to the directory you would like to install
tar xzvf epacts_v2_01.noref_binary.2012_07_06.tar.gz
Accessing help
For a list of commands available in EPACTS, type in the following commands:
'''$ epacts2/epacts help''' Usage: epacts [command] [options] Command: help Print out brief help message man Print the full documentation in man page style single Perform single variant association group Perform groupwise (burden-style) association test anno Annotate a VCF file zoom Create a locus zoom plot from epacts results meta Perform meta-analysis across multiple epacts results Visit http://genome.sph.umich.edu/wiki/EPACTS for more detailed documentation To view options for single variant testing only type in:
$ epacts2/epacts single -helpUsage: epacts single [options]
Required Options (Run epacts single -man or see wiki for more info): -vcf STR Input VCF file (tabixed and bgzipped) -ped STR Input PED file for phenotypes and covariates -out STR Prefix of output files -test STR Statistical test to use
...
Getting started in EPACTS with an example
Once installed, test out the software by running a quick example using the test data provided in the "example" directory. The example VCF and PED files are:
$ epacts2/example/1000G_exome_chr20_example_softFiltered.calls.vcf.gz $ epacts2/example/1000G_dummy_pheno.ped
Run the single variant score test on the example data using this command:
$ epacts2/epacts single \
--vcf epacts2/example/1000G_exome_chr20_example_softFiltered.calls.vcf.gz \
--ped epacts2/example/1000G_dummy_pheno.ped \
--min-maf 0.001 --chr 20 --pheno DISEASE --cov AGE --cov SEX --test b.score --anno \
--out {OUTPUT_DIR}/test --run 2 &
This command will run the single variant test on the input VCF and PED files, with a minimum MAF threshold of 0.001. The phenotype is "DISEASE" and we are adjusting the analysis with covariates AGE and SEX. The output file directory prefix is {OUTPUT_DIR}/test. Finally, EPACTS will run the analysis in parallel on 2 CPUs.
Expected output
EPACTS produces a number of files and plots.
1. test.epacts.gz contains all the association results.
> head test.epacts #CHROM BEGIN END MARKER_ID NS AC CALLRATE MAF PVALUE SCORE 20 68303 68303 20:68303_A/G_Upstream:DEFB125 266 1 1 0.0018797 NA NA 20 68319 68319 20:68319_C/A_Upstream:DEFB125 266 0 1 0 NA NA 20 68396 68396 20:68396_C/T_Nonsynonymous:DEFB125 266 1 1 0.0018797 NA NA 20 76635 76635 20:76635_A/T_Intron:DEFB125 266 0 1 0 NA NA 20 76689 76689 20:76689_T/C_Synonymous:DEFB125 266 0 1 0 NA NA 20 76690 76690 20:76690_T/C_Nonsynonymous:DEFB125 266 1 1 0.0018797 NA NA 20 76700 76700 20:76700_G/A_Nonsynonymous:DEFB125 266 0 1 0 NA NA 20 76726 76726 20:76726_C/G_Nonsynonymous:DEFB125 266 0 1 0 NA NA 20 76771 76771 20:76771_C/T_Nonsynonymous:DEFB125 266 3 1 0.0056391 0.68484 0.40587
2. test.epacts.top5000 contains the top 5000 associated variants ordered by p-value.
$ head out/test.single.b.score.epacts.top5000 #CHROM BEGIN END MARKER_ID NS AC CALLRATE MAF PVALUE SCORE 20 1610894 1610894 20:1610894_G/A_Synonymous:SIRPG 266 136 1 0.25564 0.0001097 3.8681 20 4162411 4162411 20:4162411_T/C_Intron:SMOX 266 204 1 0.38346 0.00055585 -3.4523 20 34061918 34061918 20:34061918_T/C_Intron:CEP250 266 39 1 0.073308 0.0011231 3.2577 20 4155948 4155948 20:4155948_G/A_Intron:SMOX 266 215 1 0.40414 0.0020791 -3.0787 20 4680251 4680251 20:4680251_A/G_Nonsynonymous:PRNP 266 186 1 0.34962 0.0025962 3.0119 20 36668874 36668874 20:36668874_G/A_Synonymous:RPRD1B 266 96 1 0.18045 0.003031 2.9646 20 36641871 36641871 20:36641871_G/A_Synonymous:TTI1 266 10 1 0.018797 0.004308 -2.8547 20 32664926 32664926 20:32664926_G/A_Nonsynonymous:RALY 266 20 1 0.037594 0.0046365 2.8313 20 34288854 34288854 20:34288854_C/T_Utr3:ROMO1 266 28 1 0.052632 0.0047722 2.822
3. test.epacts.qq.pdf contains the Q-Q plot of test p-values (stratified by MAF)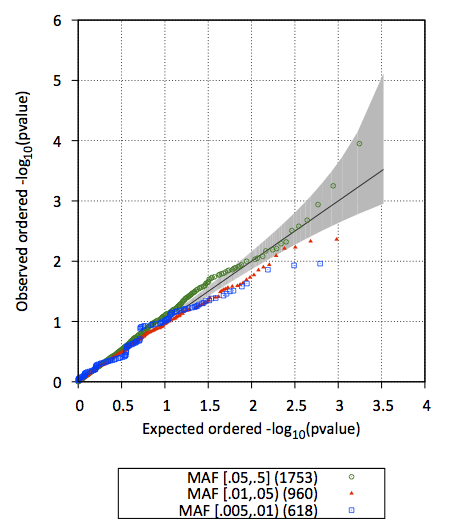
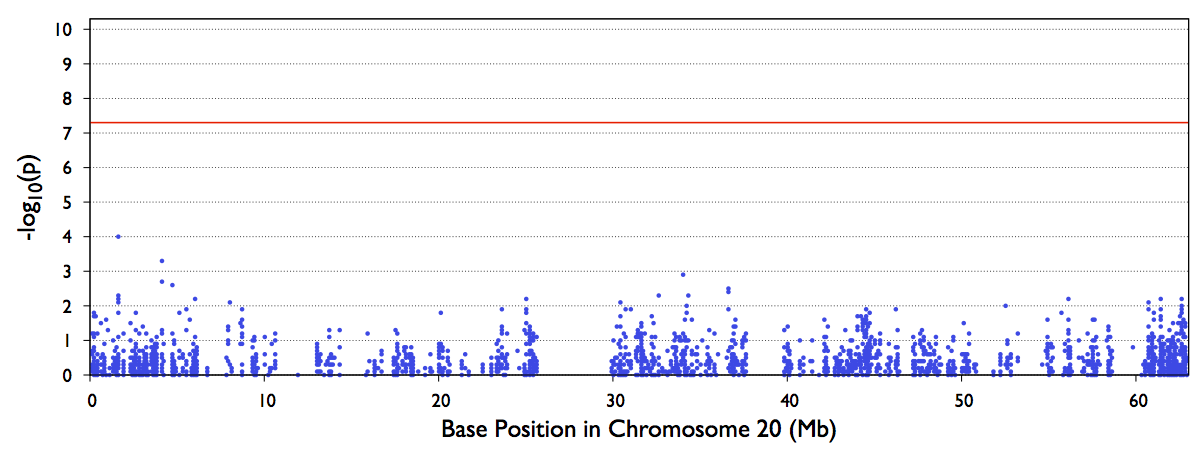
4. test.epacts.mh.pdf contains the Manhattan Plot of test p-values
The file out/test.b.score.epacts.mh.pdf will be generated for chr20 only.
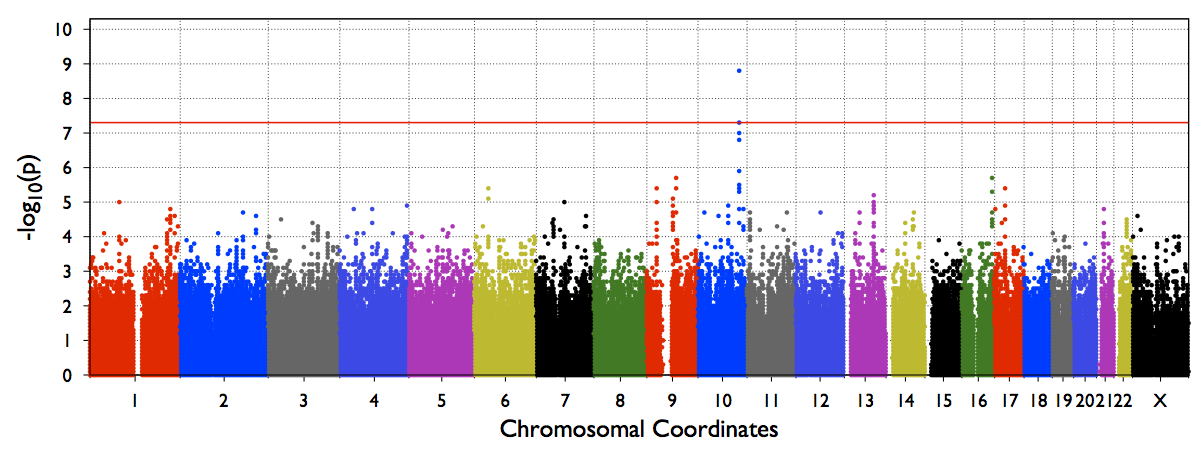
An example Genome-wide manhattan plot (from a genome-wide run) will look like below
...