Tutorial: GotCloud
GotCloud Tutorial
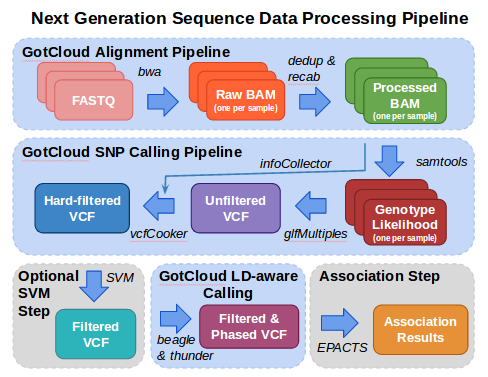
In this tutorial, we illustrate some of the essential steps in the analysis of next generation sequence data.
Analysis starts with FASTQ files, the typical format provided from your sequencing center containing the sequence & base quality information for your data.
The fastq files are processed using the alignment pipeline which finds the most likely genomic location for each read and stores that information in a BAM (Binary Sequence Alignment/Map format) file. In addition to the sequence and base quality information contained in FASTQ files, a BAM file also contains the genomic location and some additional information about the mapping. As part of the alignment pipeline, the base qualities are adjusted to more accurately reflect the likelihood that the base is correct.
The variant calling pipeline processes the BAMs file produced by the alignment pipeline, generating an initial list of polymorphic sites and genotypes stored in a VCF (Variant Call Format) file and then uses haplotype information to refine these genotypes in an updated VCF file.
This tutorial then demonstrates how EPACTS (Efficient and Parallelizable Association Container Toolbox) can be used to perform statistical tests to identify genome-wide association from sequence data.
STEP 1 : Setup GotCloud
Step 1a: Setup Environment
To facilitate running this tutorial, set environment variables:
- GCHOME to the path to the base directory of your gotcloud instalation
- GCDATA to the path to the base directory of the example data instalation
In the instructions below, we
First check your shell using:
ps -p $$ -ocomm=
For bash, sh:
export GCHOME=~/gotcloud export GCDATA=~/gotcloudExample export GCOUT=~/gotcloudTutorial
For csh, tcsh:
setenv GCHOME ~/gotcloud setenv GCDATA ~/gotcloudExample setenv GCOUT ~/gotcloudTutorial
Step 1b: Install GotCloud
In order to run this tutorial, you need to make sure you have GotCloud is installed on your system.
If you have root and would like to install gotcloud on your system, follow: root access installation instructions
Otherwise, you can install it in your own directory:
- Create & Change directory to where you want to install gotcloud
- Download the gotcloud tar from the ftp site.
- Extract the tar
- Build the source
mkdir -p $GCHOME; cd $GCHOME wget ftp://share.sph.umich.edu/gotcloud/gotcloud.tar # Download tar xvf gotcloud.tar --strip 1 # Extract cd $GCHOME/src; make # Build source
Step 1c: Install Example Dataset
Our dataset consists of 60 individuals from GBR sequenced by the 1000 Genomes Project. These individuals have been sequenced to an average depth of about 4x.
To conserve time and disk-space, our analysis will focus on a small region on chromosome 20, 42900000 - 43200000. We will first map the reads for two individuals (HG00096, HG00100).
The 2nd step is to use the mapped reads for 60 individuals to generate a list of polymorphic sites and estimate accurate genotypes at each of these sites.
The example dataset we'll be using is available at: ftp://share.sph.umich.edu/gotcloud/gotcloudExample.tar
- Create & Change directory to where you want to install the Tutorail data
- Download the dataset tar from the ftp site
- Extract the tar
mkdir -p $GCDATA; cd $GCDATA wget ftp://share.sph.umich.edu/gotcloud/gotcloudExample.tar # Download tar xvf gotcloudExample.tar --strip 1 # Extract
STEP 2 : Run GotCloud Alignment Pipeline
The first step in processing next generation sequence data is mapping the reads to the reference genome, generating per sample BAM files.
The alignment pipeline has multiple built-in steps to generate BAMs:
- Align the fastqs to the reference genome
- handles both single & paired end
- Merge the results from multiple fastqs into 1 file per sample
- Mark Duplicate Reads are marked
- Recalibrate Base Qualities
This processing results in 1 BAM file per sample.
The alignment pipeline also includes Quality Control (QC) steps:
- Visualization of various quality measures (QPLOT)
- Screen for sample contamination & swap (VerifyBamID)
Run the alignment pipeline (the example aligns 2 samples) :
$GCHOME/gotcloud align --conf $GCDATA/GBR60align.conf --outdir $GCOUT
Upon successful completion of the alignment pipeline (about 1-2 minutes), you will see the following message:
Processing finished in nn secs with no errors reported
The final BAM files produced by the alignment pipeline are:
ls $GCOUT/alignment.recal/*.recal.bam $GCOUT/alignment.recal/*.recal.bam.bai
Index files (.bai) for these BAMs are also in that directory.
The QC files for verifyBamID are:
ls $GCOUT/QCFiles/*.genoCheck.self* $GCOUT/QCFiles/*.genoCheck.depth*
Understanding VerifyBamID output
The QC files for qplot are:
ls $GCOUT/QCFiles/*.qplot.R $GCOUT/QCFiles/*.qplot.stats
STEP 3 : Run GotCloud Variant Calling Pipeline
The next step is to analyze BAM files by calling SNPs and generating a VCF file containing the results.
The variant calling pipeline has multiple built-in steps to generate BAMs:
- Filter out reads with low mapping quality
- Per Base Alignment Quality Adjustment (BAQ)
- Resolve overlapping paired end reads
- Generate genotype likelihood files
- Perform variant calling
- Extract features from variant sites
- Perform variant filtering
This processing results in a single set of variant sites for all samples.
Run the variant calling pipeline:
$GCHOME/gotcloud snpcall --conf GBR60vc.conf --outdir $GCOUT --numjobs 2 --region 20:42900000-43200000
Upon successful completion of the variant calling pipeline (about 3-4 minutes), you will see the following message:
Commands finished in nnn secs with no errors reported
The final VCF produced by the variant calling pipeline containing only the variants that passed all filters is:
ls $GCOUT/split/chr20/chr20.filtered.PASS.vcf.gz
The VCF including the filtered sites with the filters marked in the Filter field (or "PASS" if the site was not filtered) is:
ls $GCOUT/vcfs/chr20/chr20.filtered.vcf.gz
STEP 4 : Run GotCloud Genotype Refinement Pipeline
The next step is to perform genotype refinement using linkage disequilibrium information using Beagle & ThunderVCF.
Run the LD-aware genotype refinement pipeline:
$GCHOME/gotcloud ldrefine --conf GBR60vc.conf --outdir $GCOUT --numjobs 2
Upon successful completion of this pipeline, you will see the following message:
Commands finished in nnn secs with no errors reported
The output from the beagle step of the genotype refinement pipeline is found in:
ls $GCOUT/beagle/chr20/chr20.filtered.PASS.beagled.vcf.gz $GCOUT/beagle/chr20/chr20.filtered.PASS.beagled.vcf.gz.tbi
The output from the thunderVcf (final step) of the genotype refinement pipeline is found in:
ls $GCOUT/thunder/chr20/GBR/chr20.filtered.PASS.beagled.GBR.thunder.vcf.gz $GCOUT/thunder/chr20/GBR/chr20.filtered.PASS.beagled.GBR.thunder.vcf.gz.tbi
STEP 5 : Run GotCloud Association Analysis Pipeline
/net/fantasia/home/hmkang/bin/epacts/bin/epacts single --vcf $GCOUT/vcfs/chr20/chr20.filtered.vcf.gz --ped $GCDATA/test.GBR60.ped --out EPACTS_TEST --test q.linear --run 1 --top 1 --chr 20
Tutorial Inputs
Alignment Pipeline
The inputs to the tutorial alignment pipeline are:
Alignment Configuration File
The configuration file contains KEY = VALUE settings that override defaults and set specific values for the given run.
INDEX_FILE = GBR60fastq.index ############ # References REF_DIR = chr20Ref AS = NCBI37 FA_REF = $(REF_DIR)/human_g1k_v37_chr20.fa DBSNP_VCF = $(REF_DIR)/dbsnp135_chr20.vcf.gz HM3_VCF = $(REF_DIR)/hapmap_3.3.b37.sites.chr20.vcf.gz
This configuration file sets:
- INDEX_FILE - file containing the fastqs to be processed as well as the read group information for these fastqs.
- Reference Information:
- AS - assembly value to put in the BAM
- FA_REF - the reference file (.fa extension), the additional files should be at the same location:
- human_g1k_v37_chr20-bs.umfa
- human_g1k_v37_chr20.dict
- human_g1k_v37_chr20.fa
- human_g1k_v37_chr20.fa.amb
- human_g1k_v37_chr20.fa.ann
- human_g1k_v37_chr20.fa.bwt
- human_g1k_v37_chr20.fa.fai
- human_g1k_v37_chr20.fa.GCcontent
- human_g1k_v37_chr20.fa.pac
- human_g1k_v37_chr20.fa.rbwt
- human_g1k_v37_chr20.fa.rpac
- human_g1k_v37_chr20.fa.rsa
- human_g1k_v37_chr20.fa.sa
- DBSNP_VCF - a vcf containing the dbsnp positions
- HM3_VCF - hapmap vcf
For running your own test, update the INDEX_FILE to point to your index file and the reference values to point to your references.
This example uses reference files that are chr20 only in order to speed processing of the tutorial data. If you are using the default references, you may just need to update REF_DIR to the directory where they are installed. Full Reference files can be downloaded from GotCloudReference.
It is recommended that you use absolute paths. (This example does not use absolute paths in order to be flexible to where the data is installed, but using relative paths require it to be run from the correct directory.)
Alignment Output Directory
This setting tells the pipeline what directory to write the output files into.
The output directory will be created if necessary and will contain the following Directories/files:
- bams - directory containing the final bams/bai files
- HG00096.OK - indicates that this sample completed alignment processing
- HG00100.OK - indicates that this sample completed alignment processing
- failLogs - directory containing logs from steps that failed
- Makefiles - directory containing the makefiles for each sample
- QCFiles - directory containing the QC Results
- tmp - directory containing temporary alignment files
Modifying the Tutorial Inputs to Run Your Own Data
Download the whole genome resource files
If you want to analyze sequence data beyond chr20, you will first need to download the full resource files from TBA
Index file
There are four fastq files in {ROOT_DIR}/test/align/fastq/Sample_1 and four fastq files in {ROOT_DIR}/test/align/fastq/Sample_2, both in paired-end format. Normally, we would need to build an index file for these files. Conveniently, an index file (indexFile.txt) already exists for the automatic test samples. It can be found in {ROOT_DIR}/test/align/, and contains the following information in tab-delimited format:
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM Sample1 fastq/Sample_1/File1_R1.fastq.gz fastq/Sample_1/File1_R2.fastq.gz RGID1 SampleID1 Lib1 UM ILLUMINA Sample1 fastq/Sample_1/File2_R1.fastq.gz fastq/Sample_1/File2_R2.fastq.gz RGID1a SampleID1 Lib1 UM ILLUMINA Sample2 fastq/Sample_2/File1_R1.fastq.gz fastq/Sample_2/File1_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA Sample2 fastq/Sample_2/File2_R1.fastq.gz fastq/Sample_2/File2_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA
If you are in the {ROOT_DIR}/test/align directory, you can use this file as-is. If you prefer, you can create a new index file and change the MERGE_NAME, RGID, SAMPLE, LIBRARY, CENTER, or PLATFORM values. It is recommended that you do not modify existing files in {ROOT_DIR}/test/align.
If you want to run this example from a different directory, make sure the FASTQ1 and FASTQ2 paths are correct. That is, each of the FASTQ1 and FASTQ2 entry in the index file should look like the following:
{ROOT_DIR}/test/align/fastq/Sample_1/File1_R1.fastq.gz
Alternately, if you want to run this example from a different directory, but do not want to edit the index file, you can create a relative path to the test fastq files so their path agrees with that listed in the index file:
ln -s {ROOT_DIR}/test/align/fastq fastq
This will create a symbolic link to the test fastq directory from your current directory.
(More information about: the index file.)
Configuration file
Similar to the index file, a configuration file (test.conf) already exists for the automatic test samples. It contains the following information:
INDEX_FILE = indexFile.txt ############ # References REF_DIR = $(PIPELINE_DIR)/test/align/chr20Ref AS = NCBI37 FA_REF = $(REF_DIR)/human_g1k_v37_chr20.fa DBSNP_VCF = $(REF_DIR)/dbsnp.b130.ncbi37.chr20.vcf.gz PLINK = $(REF_DIR)/hapmap_3.3.b37.chr20
If you are in the {ROOT_DIR}/test/align directory, you can use this file as-is. If you are using a different index file, make sure your index file is named correctly in the first line. If you are not running this from {ROOT_DIR}/test/align, make sure your configuration and index files are in the same directory.
(More information about: reference files, optional configurable settings, or command-line options.)
Running the alignment pipeline
You are now ready to run the alignment pipeline.
To run the alignment pipeline, enter the following command:
{ROOT_DIR}/bin/gen_biopipeline.pl -conf test.conf -out_dir {OUT_DIR}
where {OUT_DIR} is the directory in which you wish to store the resulting BAM files (for example, ~/out).
If everything went well, you will see the following messages:
Created {OUT_DIR}/Makefiles/biopipe_Sample2.Makefile
Created {OUT_DIR}/Makefiles/biopipe_Sample1.Makefile
---------------------------------------------------------------------
Submitted 2 commands
Waiting for commands to complete... . . Commands finished in 33 secs with no errors reported
The aligned BAM files are found in {OUT_DIR}/alignment.recal/
Analyzing a Sample
Using UMAKE, you can analyze BAM files by calling SNPs, and generate a VCF file containing the results. Once again, we can analyze BAM files used in the automatic test. For this example, we have 60 BAM files, which can be found in {ROOT_DIR}/test/umake/bams. These contain sequence information for a targeted region in chromosome 20.
In addition to the BAM files, you will need three files to run UMAKE: an index file, a configuration file, and a bed file (needed to analyze BAM files from targeted/exome sequencing).
Index file
First, you need a list of all the BAM files to be analyzed. Conveniently, the a test index file (umake_test.index) already exists in {ROOT_DIR}/test/umake/. It contains the following information:
NA12272 ALL bams/NA12272.mapped.ILLUMINA.bwa.CEU.low_coverage.20101123.chrom20.20000001.20300000.bam NA12004 ALL bams/NA12004.mapped.ILLUMINA.bwa.CEU.low_coverage.20101123.chrom20.20000001.20300000.bam ... NA12874 ALL bams/NA12874.mapped.LS454.ssaha2.CEU.low_coverage.20101123.chrom20.20000001.20300000.bam
You can use this file directly if you change your current directory to {ROOT_DIR}/test/umake/.
Alternately, if you want to copy and use this index file to a different directory, you can create a symbolic link to the bams folder as follows:
ln -s {ROOT_DIR}/test/umake/bams bams
(More information about: the index file.)
BED file
This file contains a single line:
chr20 20000050 20300000
You can copy this to the current directory and use it as-is.
(More information about: targeted/exome sequencing settings.)
Configuration file
A configuration file (umake_test.conf) already exists in {ROOT_DIR}/test/umake/. It contains the following information:
CHRS = 20 TEST_ROOT = $(UMAKE_ROOT)/test/umake BAM_INDEX = $(TEST_ROOT)/umake_test.index OUT_PREFIX = umake_test REF_ROOT = $(TEST_ROOT)/ref # REF = $(REF_ROOT)/karma.ref/human.g1k.v37.chr20.fa INDEL_PREFIX = $(REF_ROOT)/indels/1kg.pilot_release.merged.indels.sites.hg19 DBSNP_PREFIX = $(REF_ROOT)/dbSNP/dbsnp_135_b37.rod HM3_PREFIX = $(REF_ROOT)/HapMap3/hapmap3_r3_b37_fwd.consensus.qc.poly # RUN_INDEX = TRUE # create BAM index file RUN_PILEUP = TRUE # create GLF file from BAM RUN_GLFMULTIPLES = TRUE # create unfiltered SNP calls RUN_VCFPILEUP = TRUE # create PVCF files using vcfPileup and run infoCollector RUN_FILTER = TRUE # filter SNPs using vcfCooker RUN_SPLIT = TRUE # split SNPs into chunks for genotype refinement RUN_BEAGLE = FALSE # BEAGLE - MUST SET AFTER FINISHING PREVIOUS STEPS RUN_SUBSET = FALSE # SUBSET FOR THUNDER - MAY BE SET WITH BEAGLE STEP TOGETHER RUN_THUNDER = FALSE # THUNDER - MUST SET AFTER FINISHING PREVIOUS STEPS ############################################################################### WRITE_TARGET_LOCI = TRUE # FOR TARGETED SEQUENCING ONLY -- Write loci file when performing pileup UNIFORM_TARGET_BED = $(TEST_ROOT)/umake_test.bed # Targeted sequencing : When all individuals has the same target. Otherwise, comment it out OFFSET_OFF_TARGET = 50 # Extend target by given # of bases MULTIPLE_TARGET_MAP = # Target per individual : Each line contains [SM_ID] [TARGET_BED] TARGET_DIR = target # Directory to store target information SAMTOOLS_VIEW_TARGET_ONLY = TRUE # When performing samtools view, exclude off-target regions (may make command line too long)
If you are running this from a different directory, you will want to change some of the lines as follows:
BAM_INDEX = {CURRENT_DIR}/umake_test.index
UNIFORM_TARGET_BED = {CURRENT_DIR}/umake_test.bed
where {CURRENT_DIR} is the absolute path to the directory that contains the index and bed files.
An additional option can be added in the configuration file:
OUT_DIR = {OUT_DIR}
where {OUT_DIR} is the name of directory in which you want the output to be stored. If you do not specify this in the configuration file, you will need to add an extra parameter when you run UMAKE in the next step.
(More information about: the configuration file, reference files.)
Running UMAKE
If you added an OUT_DIR line to the configuration file, you can run UMAKE with the following command:
{ROOT_DIR}/bin/umake.pl --conf umake_test.conf --snpcall --numjobs 2
If you have not added an OUT_DIR line to the configuration file, you can specify the output directory directly with the following command:
{ROOT_DIR}/bin/umake.pl --conf umake_test.conf --outdir {OUT_DIR} --snpcall --numjobs 2
where {OUT_DIR} is the directory in which you want the output to be stored.
Either command will perform SNP calling on the test samples. If you find the resulting VCF files located in {OUT_DIR}/vcfs/chr20, then you have successfully called the SNPs from the test BAM files.