Difference between revisions of "Understanding vcf-summary output"
(Created page with "== What is vcf-summary? == <code>vcf-summary</code> is a utility included in GotCloud that helps evaluate the quality of SNP calls. Because GotCloud will automaticall...") |
|||
| Line 22: | Line 22: | ||
In addition, multi-allelic or duplicated SNPs are counted separately at the very bottom. | In addition, multi-allelic or duplicated SNPs are counted separately at the very bottom. | ||
| + | |||
| + | == Understanding the columns of vcf-summary output == | ||
| + | |||
| + | vcf-summary output can be a little bit dense to digest at the first time, but contains useful information about the quality of SNP calls. | ||
| + | |||
| + | First, define transitions and transversions.. | ||
| + | * '''Transition''' (Ts or Ti) refers to the bi-allelic SNPs where two alleles are A <-> G or C <-> T. In true SNPs, transitions more frequently occurs than transversions. | ||
| + | * '''Transversion''' (Tv) refers to the bi-allelic SNPs that are not transitions. | ||
| + | |||
| + | Each column of vcf-summary output shows.. | ||
| + | |||
| + | * '''FILTER''' shows the groups of SNPs represented by FILTER columns in the VCF file. See previous section to know how SNPs are grouped. | ||
| + | * '''#SNPs''' shows the total number of SNPs that belong to each FILTER category. | ||
| + | * '''#dbSNPs''' shows the number of SNPs that appear in the dbSNP. The version of dbSNP can be different based on the GotCloud version (e.g. 129, 135, 137) | ||
| + | * '''%dbSNPs''' are simply [#dbSNPs] / [#SNPs], indicating the fraction of variants ''known'' by dbSNP. The rest of them will be considered as ''novel'' | ||
| + | * '''%CpG Known''' are the fraction of known (in dbSNP) SNPs that are located at CpG sites, which are enriched for transitions. | ||
| + | * '''%CpG Novel''' are the fraction of novel (not in dbSNP) SNPs that are located at CpG sites, which are enriched for transitions. | ||
| + | * '''Known Ts/Tv''' represents Ts/Tv ratio among known (in dbSNP) SNPs. | ||
| + | * '''Novel Ts/Tv''' represents Ts/Tv ratio among novel (not in dbSNP) SNPs. | ||
| + | * '''nCpG-K Ts/Tv''' represents Ts/Tv ratio among known (in dbSNPs) SNPs, when only focusing on non-CpG sites. Ts/Tv becomes more stable when excluding CpG sites. | ||
| + | * '''nCpG-N Ts/Tv''' represents Ts/Tv ratio among novel (in dbSNPs) SNPs, when only focusing on non-CpG sites. | ||
| + | * '''%HM3 sens''' indicate the percent of HapMap3 SNPs rediscovered among all HapMap SNPs ( [# HapMap3-overlapping SNPs] / [# all HapMap 3 SNPs] ) | ||
| + | * '''%HM3/SNP''' indicate the percent of HapMap3 SNPs among all SNPs in the call set ( [# HapMap3-overlapping SNPs] / [# all SNPs in the call set] ) | ||
| + | |||
| + | == What should we expect from good SNP call sets? == | ||
| + | |||
| + | If the SNP filtering was effective, you will expect the following properties in "PASS" category, but not in the other categories. | ||
| + | |||
| + | * Metrics between '''Known''' and '''Novel''' categories should be similar. This indicates that novel SNPs show as good qualities as known SNPs, which are enriched for true variants. | ||
| + | * Expected '''Ts/Tv''' values for whole genome call set are around 2.2. '''non-CpG Ts/Tv''' are usually ranged around 1.8 | ||
| + | * For exome call sets, Ts/Tv are expected to be higher (around 3). Synonymous SNPs will have higher Ts/Tv (around 5.5) than non-synonymous SNPs (around 2.2). | ||
| + | * Because of natural selection, in exomes, we expect higher Ts/Tv in known SNPs than novel SNPs. This is because novel SNPs are more likely to be non-synonymous SNPs than synonymous SNPs. Please refer to [[SNP_Call_Set_Properties]] for additional information. | ||
| + | * '''%dbSNP''' should be usually higher for '''PASS''' categories than others, indicating that known SNPs are more likely to pass filters. However, because dbSNP also contain false positive variants, we don't necessarily expect that '''%dbSNP''' go to zero even if the filtering works perfectly. | ||
| + | * Because HM3 consist of common SNPs, this number should be high when the sample size is large. Typically, if the sample contains only Europeans, the expected values of '''%HM3 sens''' are around 90%. If the sample contains African ancestry, the number will be close to 95% of higher. | ||
| + | * Note that some callers uses the expected difference between Ts and Tv in their calling model as prior. In that case, Ts/Tv could be biased by the prior, and Ts/Tv may not be a good indicator of SNP qualities. | ||
Revision as of 23:29, 16 June 2014
What is vcf-summary?
vcf-summary is a utility included in GotCloud that helps evaluate the quality of SNP calls. Because GotCloud will automatically run vcf-summary, detailed instructions on the usage of the program is not currently documented.
Example output from vcf-summary
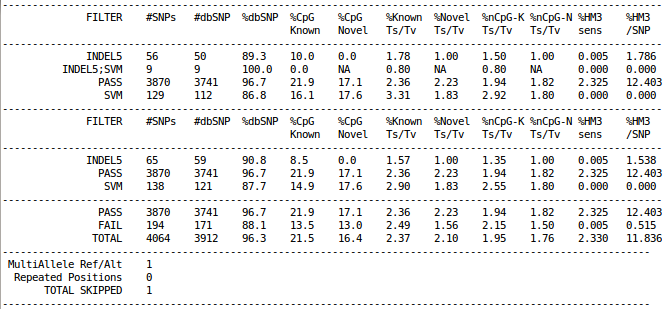
If OUT is an environment variable, you may see output file from GotCloud similar to the following example.
cat ${OUT}/vcfs/chr22/chr22.filtered.sites.vcf.summary
The example above is obtained from the results of GotCloud within a very small (1Mb) region in chr22 across ~60 1000 Genomes samples.
Rows of vcf-summary output has three sections
As shown in the example figure above, typical vcf-summary output primarily consists of the following three sections.
- In the first part, each SNP is counted only once, grouped by the contents of FILTER column.
- In the second part, each SNP may be counted multiple times, if the SNP failed multiple filters (e.g. INDEL5 filter and SVM filter).
- In the last part, each SNP is counted only once, grouped by SNPs with "PASS" in the FILTER column versus everything else.
In addition, multi-allelic or duplicated SNPs are counted separately at the very bottom.
Understanding the columns of vcf-summary output
vcf-summary output can be a little bit dense to digest at the first time, but contains useful information about the quality of SNP calls.
First, define transitions and transversions..
- Transition (Ts or Ti) refers to the bi-allelic SNPs where two alleles are A <-> G or C <-> T. In true SNPs, transitions more frequently occurs than transversions.
- Transversion (Tv) refers to the bi-allelic SNPs that are not transitions.
Each column of vcf-summary output shows..
- FILTER shows the groups of SNPs represented by FILTER columns in the VCF file. See previous section to know how SNPs are grouped.
- #SNPs shows the total number of SNPs that belong to each FILTER category.
- #dbSNPs shows the number of SNPs that appear in the dbSNP. The version of dbSNP can be different based on the GotCloud version (e.g. 129, 135, 137)
- %dbSNPs are simply [#dbSNPs] / [#SNPs], indicating the fraction of variants known by dbSNP. The rest of them will be considered as novel
- %CpG Known are the fraction of known (in dbSNP) SNPs that are located at CpG sites, which are enriched for transitions.
- %CpG Novel are the fraction of novel (not in dbSNP) SNPs that are located at CpG sites, which are enriched for transitions.
- Known Ts/Tv represents Ts/Tv ratio among known (in dbSNP) SNPs.
- Novel Ts/Tv represents Ts/Tv ratio among novel (not in dbSNP) SNPs.
- nCpG-K Ts/Tv represents Ts/Tv ratio among known (in dbSNPs) SNPs, when only focusing on non-CpG sites. Ts/Tv becomes more stable when excluding CpG sites.
- nCpG-N Ts/Tv represents Ts/Tv ratio among novel (in dbSNPs) SNPs, when only focusing on non-CpG sites.
- %HM3 sens indicate the percent of HapMap3 SNPs rediscovered among all HapMap SNPs ( [# HapMap3-overlapping SNPs] / [# all HapMap 3 SNPs] )
- %HM3/SNP indicate the percent of HapMap3 SNPs among all SNPs in the call set ( [# HapMap3-overlapping SNPs] / [# all SNPs in the call set] )
What should we expect from good SNP call sets?
If the SNP filtering was effective, you will expect the following properties in "PASS" category, but not in the other categories.
- Metrics between Known and Novel categories should be similar. This indicates that novel SNPs show as good qualities as known SNPs, which are enriched for true variants.
- Expected Ts/Tv values for whole genome call set are around 2.2. non-CpG Ts/Tv are usually ranged around 1.8
- For exome call sets, Ts/Tv are expected to be higher (around 3). Synonymous SNPs will have higher Ts/Tv (around 5.5) than non-synonymous SNPs (around 2.2).
- Because of natural selection, in exomes, we expect higher Ts/Tv in known SNPs than novel SNPs. This is because novel SNPs are more likely to be non-synonymous SNPs than synonymous SNPs. Please refer to SNP_Call_Set_Properties for additional information.
- %dbSNP should be usually higher for PASS categories than others, indicating that known SNPs are more likely to pass filters. However, because dbSNP also contain false positive variants, we don't necessarily expect that %dbSNP go to zero even if the filtering works perfectly.
* Because HM3 consist of common SNPs, this number should be high when the sample size is large. Typically, if the sample contains only Europeans, the expected values of %HM3 sens are around 90%. If the sample contains African ancestry, the number will be close to 95% of higher.
- Note that some callers uses the expected difference between Ts and Tv in their calling model as prior. In that case, Ts/Tv could be biased by the prior, and Ts/Tv may not be a good indicator of SNP qualities.