Difference between revisions of "Variant Normalization"
| (45 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= Introduction = | = Introduction = | ||
| − | The Variant Call Format (VCF) is a flexible file format specification that allows us to represent many different variant types ranging from SNPs, | + | The Variant Call Format (VCF) is a flexible file format specification that allows us to represent many different variant types ranging from SNPs, indels to copy number variations. However, variant representation in VCF is non-unique for variants that have explicitly expressed reference and alternate sequences. A failure to recognize this will frequently result in inaccurate analyses. |
| − | On this wiki page, we describe a variant normalization procedure that is well defined for biallelic as well as multiallelic variants | + | On this wiki page, we describe a variant normalization procedure that is well defined for biallelic as well as multiallelic variants. We then provide a formal proof the procedure's correctness. |
| − | = | + | = Definition = |
The normalization of a variant representation in VCF consists of two parts: parsimony and left alignment pertaining to the nature of a variant's length and position respectively. | The normalization of a variant representation in VCF consists of two parts: parsimony and left alignment pertaining to the nature of a variant's length and position respectively. | ||
| Line 11: | Line 11: | ||
== Parsimony == | == Parsimony == | ||
| − | + | In the context of variant representation, parsimony means representing a variant in as few nucleotides as possible without reducing the length of any allele to 0. It is a property describing the nature of the length of a variant's alleles and is defined as follows: | |
| − | A variant is parsimonious if it is represented in as few nucleotides as possible | + | A variant is parsimonious if and only if it is represented in as few nucleotides as possible |
without an allele of length 0. | without an allele of length 0. | ||
| + | |||
| + | Also, | ||
| + | |||
| + | A variant has superfluous nucleotides on its left side if the leftmost nucleotide of each variant is of the same type | ||
| + | and the removal of the nucleotide from each allele will not result in an empty allele. | ||
Taking the example below, the Multi Nucleotide Polymorphism (MNP) is represented superfluously for the first 3 representations and parsimoniously for the 4th representation. When a variant has superfluous nucleotides on the left side, it is defined as not being left parsimonious as there is a need to left trim. The concept is symmetric for right parsimony and trimming. Parsimony applies to Indels too which we shall demonstrate in the left alignment section. | Taking the example below, the Multi Nucleotide Polymorphism (MNP) is represented superfluously for the first 3 representations and parsimoniously for the 4th representation. When a variant has superfluous nucleotides on the left side, it is defined as not being left parsimonious as there is a need to left trim. The concept is symmetric for right parsimony and trimming. Parsimony applies to Indels too which we shall demonstrate in the left alignment section. | ||
| Line 32: | Line 37: | ||
Left aligning a variant means shifting the start position of that variant to the left till it is no longer possible to do so. It is a concept associated with insertion and deletion variants and describes specifically the nature of a position of a variant as opposed to its length. In order to further differentiate left alignment from simply left padding a variant, the definition is as follows: | Left aligning a variant means shifting the start position of that variant to the left till it is no longer possible to do so. It is a concept associated with insertion and deletion variants and describes specifically the nature of a position of a variant as opposed to its length. In order to further differentiate left alignment from simply left padding a variant, the definition is as follows: | ||
| − | A variant is left aligned if it is no longer possible to shift its position | + | A variant is left aligned if and only if it is no longer possible to shift its position |
to the left while keeping the length of all its alleles constant. | to the left while keeping the length of all its alleles constant. | ||
| − | The following figure shows examples of an unnormalized short tandem repeat which is a special class of indels. The | + | The following figure shows examples of an unnormalized short tandem repeat which is a special class of indels. The color of the text is synchronized to the variant that is described in the figure. |
| − | * <span style="color:#ff0000">The representation of variants in a VCF file requires that no alleles in the REF and ALT field are represented with an empty string (empty allele). The red indel | + | * <span style="color:#ff0000">The representation of variants in a VCF file requires that no alleles in the REF and ALT field are represented with an empty string (empty allele). The red indel has an illegal VCF representation.</span> |
* <span style="color:#008000"> The green variant is not left aligned as you can prefix an A nucleotide on the left side of the variant's alleles and truncate the C on the right side of the variant's alleles. It is however parsimonious. </span> | * <span style="color:#008000"> The green variant is not left aligned as you can prefix an A nucleotide on the left side of the variant's alleles and truncate the C on the right side of the variant's alleles. It is however parsimonious. </span> | ||
* <span style="color:#F9A908"> The orange variant is left aligned but is not right parsimonious.</span> | * <span style="color:#F9A908"> The orange variant is left aligned but is not right parsimonious.</span> | ||
| Line 46: | Line 51: | ||
This figure shows multiple representations of a CA tandem repeat. The left shows five possible representations differentiated by color. The right shows the corresponding representation in VCF. The last representation represents the left aligned and parsimonious representation of the STR. | This figure shows multiple representations of a CA tandem repeat. The left shows five possible representations differentiated by color. The right shows the corresponding representation in VCF. The last representation represents the left aligned and parsimonious representation of the STR. | ||
| − | + | = When is a variant normalized? = | |
A variant is normalized if and only if it is parsimonious and left aligned. | A variant is normalized if and only if it is parsimonious and left aligned. | ||
| Line 59: | Line 64: | ||
For the => direction, we prove the contrapositive : | For the => direction, we prove the contrapositive : | ||
| − | * If an | + | * If an indel is left aligned and right parsimonious then each allele does not end with the same type of nucleotide. |
| − | We first assume an indel is already left aligned and right parsimonious. Suppose all alleles have a length greater than 1, since the indel is right parsimonious | + | We first assume an indel is already left aligned and right parsimonious. Suppose all alleles have a length greater than 1, since the indel is right parsimonious; clearly, each allele do not end with the same type of nucleotide. Now, suppose that there exists an allele of length 1, and that all the alleles end with a particular nucleotide say 'A'. This is still considered right parsimonious as there are no superfluous nucleotides to remove without resulting in an empty allele. It is possible to extend all the alleles one position to the left by copying from a nucleotide on the reference genome, so now we have a superfluous nucleotide on the right side. Trimming off that nucleotide results in a new representation that shifts the indel to the left by one position while retaining the allele lengths of the original representation. This is left aligning the indel and thus there is a contradiction. Therefore each allele cannot end with the same type of nucleotide. |
For the <= direction : | For the <= direction : | ||
| − | * If an | + | * If an indel is not left aligned or not right parsimonious then each allele ends with the same type of nucleotide. |
| − | Suppose a variant is not left aligned, then it must be possible to extend the alleles one nucleotide to the left and remove one nucleotide from the right to | + | Suppose a variant is not left aligned, then it must be possible to extend the alleles one nucleotide to the left and remove one nucleotide from the right to ensure that all the alleles remain the same length. Thus each allele must end with the same type of nucleotide for the removal of the rightmost nucleotide to be possible. |
| − | Suppose a variant is not right parsimonious, then | + | Suppose a variant is not right parsimonious, then by definition, the rightmost nucleotide is the same for all alleles which may be removed. |
== Corollary == | == Corollary == | ||
| Line 75: | Line 80: | ||
A variant is normalized if and only if | A variant is normalized if and only if | ||
1. it has no superfluous nucleotides on the left side and | 1. it has no superfluous nucleotides on the left side and | ||
| − | 2. each allele | + | 2. each allele does not end with the same type of nucleotide. |
Proof: | Proof: | ||
| Line 82: | Line 87: | ||
:⇒ A variant is normalized if and only if it is left parsimonious and right parsimonious and left aligned. (break up parsimonious definition) <br> | :⇒ A variant is normalized if and only if it is left parsimonious and right parsimonious and left aligned. (break up parsimonious definition) <br> | ||
:⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and right parsimonious and left aligned. (definition of left parsimony) <br> | :⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and right parsimonious and left aligned. (definition of left parsimony) <br> | ||
| − | :⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and each allele | + | :⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and each allele does not end with the same type of nucleotide. (lemma 1) <br> |
| + | |||
| + | == Uniqueness == | ||
| + | |||
| + | It is important that the normalization results in a unique representation of the variant. Before we begin the proof, intuitively, accept that any representation of a variant can be | ||
| + | transformed to another representation by adding nucleotides from the reference sequence to either ends of all the alleles at the same time or removing equivalent nucleotides from the ends of | ||
| + | all the alleles at the same time. | ||
| + | |||
| − | + | Now suppose there are 2 normalized variants A and B. Suppose A is at a different position from B and B is to the right of A (without loss in generality), this is not possible because by the definition of a normalized variant, it is left aligned, | |
| + | and if they were at different positions, that means B may be left aligned to A since they represent the same variants leading to a contradiction. So A and B must be at the same position. | ||
| − | |||
| − | |||
| − | + | Now, suppose that A and B are at the same position but are of different lengths where B is longer than A (without loss in generality), this is not possible as B is then not parsimonious, so B can be trimmed to the same length as A. | |
| − | + | Thus A and B have to be at the same position and have the same length and variant normalization is unique. | |
= Implementation = | = Implementation = | ||
| Line 98: | Line 109: | ||
This is implemented in [http://genome.sph.umich.edu/wiki/Vt#Normalization vt]. | This is implemented in [http://genome.sph.umich.edu/wiki/Vt#Normalization vt]. | ||
| − | = | + | == Algorithm for Normalization == |
| − | The following table shows the number of variants normalized for an anonymous data set. | + | So now that we know how to tell if a variant is normalized, we simply need to manipulate the variant till the rightmost ends of the alleles are not the same, and apply truncation to the superfluous nucleotides on the left side of the variant to obtain a normalized variant. |
| + | The algorithm to normalize a biallelic or multiallelic variant is as follows: | ||
| + | |||
| + | [[Image:variant_normalization_algorithm.png|none|600px|Lines 1 to 8 performs the left alignment and ensures parsimonious representation on the right side. Lines 9 to 11 ensures parsimonious representation on the left side. ]] | ||
| + | |||
| + | |||
| + | Lines 1 to 8 performs the left alignment and ensures parsimonious representation on the right side. Lines 9 to 11 ensures parsimonious representation on the left side. | ||
| + | |||
| + | == Comparisons == | ||
| + | |||
| + | === 20 May 2014 === | ||
| + | |||
| + | The following table shows the number of variants normalized for an anonymous data set. This analysis was done on 20 May 2014. | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 126: | Line 149: | ||
| 0 | | 0 | ||
| 57 | | 57 | ||
| − | | 57 variants from GATK's normalization were left aligned by vt. 6 were biallelic and 51 were multiallelic. Note that 2 variants were changed by GATK but were not completely normalized. | + | | 57 variants from GATK's normalization were left aligned by vt. 6 were biallelic and 51 were multiallelic. Note that 2 variants were changed by GATK but were not completely normalized. |
|- | |- | ||
| #normalized after vt | | #normalized after vt | ||
| Line 144: | Line 167: | ||
*GATK v3.1-1-g07a4bf8 | *GATK v3.1-1-g07a4bf8 | ||
*vt normalize v0.5 | *vt normalize v0.5 | ||
| + | |||
| + | Issues have been communicated to bcftools and gatk developers on 20 May 2014. | ||
| + | |||
| + | === 22 May 2014 === | ||
| + | |||
| + | The following table shows the number of variants normalized for an anonymous data set. This analysis was done on 22 May 2014. | ||
| + | bcftools was updated. | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! scope="col"| Dataset | ||
| + | ! scope="col"| bcftools | ||
| + | ! scope="col"| gatk | ||
| + | ! scope="col"| vt | ||
| + | ! scope="col"| comments | ||
| + | |- | ||
| + | | #normalized | ||
| + | | 18849 | ||
| + | | 18794 | ||
| + | | 18849 | ||
| + | | bcftools's algorithm is the same as vt now | ||
| + | |- | ||
| + | | #normalized after bcftools | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | no variants processed by bcftools were further normalized. | ||
| + | |- | ||
| + | | #normalized after gatk | ||
| + | | 57 | ||
| + | | 0 | ||
| + | | 57 | ||
| + | | GATK's algorithm is documented to work only for biallelic simple indels. The 57 variants were either multiallelic or mixed variants (SNP adjacent to indel). 6 biallelic mixed variants that were not left aligned however turned out to be simple indels after left alignment. | ||
| + | |- | ||
| + | | #normalized after vt | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | 0 | ||
| + | | no variants processed by vt were further normalized. | ||
| + | |} | ||
| + | |||
| + | Commands used are: | ||
| + | *bcftools norm -f ref.fa in.vcf -O z > out.vcf.gz | ||
| + | *java -jar GenomeAnalysisTK.jar -T LeftAlignAndTrimVariants --trimAlleles -R ref.fa --variant in.vcf.gz -o out.vcf.gz | ||
| + | *vt normalize -r ref.vcf.gz -o out.vcf.gz | ||
| + | |||
| + | Versions are: | ||
| + | *bcftools v0.2.0-rc8-5-g0e06231 (using htslib 0.2.0-rc8-6-gd49dfa6) [updated non release development version] | ||
| + | *GATK v3.1-1-g07a4bf8 | ||
| + | *vt normalize v0.5 | ||
| + | |||
| + | = Here is an example where this normalization algorithm fails = | ||
| + | |||
| + | We distinguish the concepts of normalization and decomposition/reconstruction of variants as follows: | ||
| + | |||
| + | |||
| + | Normalization involves reducing representations of a variant to a canonical representation. Normalization can be applied to biallelic variants or multiallelic variants. The problem of normalization is solvable and there exists a unique representation that is left aligned and parsimonious. Mathematical proof is published. [http://bioinformatics.oxfordjournals.org/content/suppl/2015/02/19/btv112.DC1/VtNormApplicationNote_supp_20141113_1346.pdf] | ||
| + | |||
| + | |||
| + | Decomposition of variants involves the breaking down of a variant record into multiple records. It may be done vertically - as in multiallelics becoming biallelics or it can be done horizontally - a cluster of indels and SNPs represented as a complex variant being splitted up into several records. Horizontal decompositions in general do not have a unique solution. Similarly, reconstruction combines several variant records into a single record and can be done vertically and horizontally too. Vertical decomposition of a multiallelic variant to a set of biallelic records is a many to one function. Construction of a set of biallelic variants into a multiallelic record is not unique as you need to considered all possible permutations of the haplotypes containing your alleles. | ||
| + | |||
| + | |||
| + | If your example contains the decomposition or reconstruction of variants, then it is probable that you can find inconsistencies. | ||
| + | |||
| + | |||
| + | It is important to distinguish the difference between normalization and decomposition/reconstruction. The notion of normalization implies that a variant can be reduced to a standardized form. If you were to include decomposition and reconstruction in your notion of normalization, you are bound to find inconsistencies simply due to the inherent issues of identifiability. | ||
| + | |||
| + | |||
| + | When performing decomposition and construction, I think the following factors should be considered: | ||
| + | |||
| + | * Are your variants describing just a single individual or a population? | ||
| + | * Are the genotypes (if any) in your individual(s) phased? | ||
| + | |||
| + | Depending on the context, you will obtain different answers. | ||
| + | |||
| + | [https://github.com/atks/vt/issues/16 An example of inconsistent variant representation due to using vt normalize] | ||
| + | |||
| + | = Citation = | ||
| + | |||
| + | [http://bioinformatics.oxfordjournals.org/content/31/13/2202 Adrian Tan, Gonçalo R. Abecasis and Hyun Min Kang. (2015) Unified Representation of Genetic Variants. Bioinformatics.] | ||
| + | |||
| + | = Translations = | ||
| + | |||
| + | A mandarin translation can be found [http://www.lyon0804.com/fan-yi-variant-normalization.html here] | ||
= Maintained by = | = Maintained by = | ||
This page is maintained by [mailto:atks@umich.edu Adrian]. | This page is maintained by [mailto:atks@umich.edu Adrian]. | ||
Latest revision as of 09:55, 1 August 2016
Introduction
The Variant Call Format (VCF) is a flexible file format specification that allows us to represent many different variant types ranging from SNPs, indels to copy number variations. However, variant representation in VCF is non-unique for variants that have explicitly expressed reference and alternate sequences. A failure to recognize this will frequently result in inaccurate analyses.
On this wiki page, we describe a variant normalization procedure that is well defined for biallelic as well as multiallelic variants. We then provide a formal proof the procedure's correctness.
Definition
The normalization of a variant representation in VCF consists of two parts: parsimony and left alignment pertaining to the nature of a variant's length and position respectively.
Parsimony
In the context of variant representation, parsimony means representing a variant in as few nucleotides as possible without reducing the length of any allele to 0. It is a property describing the nature of the length of a variant's alleles and is defined as follows:
A variant is parsimonious if and only if it is represented in as few nucleotides as possible without an allele of length 0.
Also,
A variant has superfluous nucleotides on its left side if the leftmost nucleotide of each variant is of the same type and the removal of the nucleotide from each allele will not result in an empty allele.
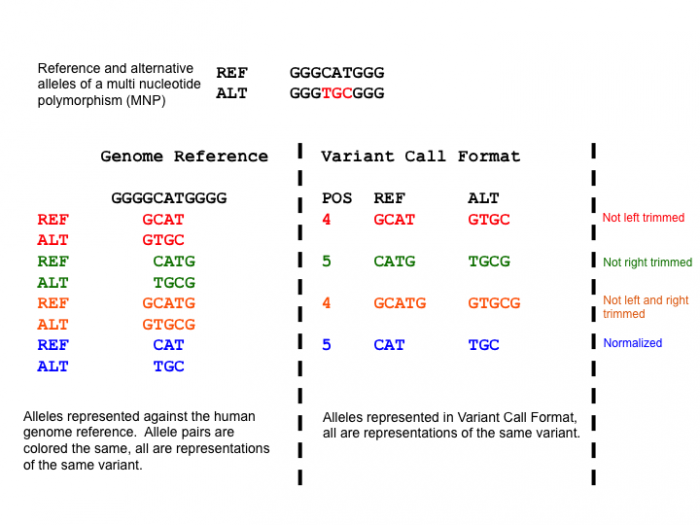
Taking the example below, the Multi Nucleotide Polymorphism (MNP) is represented superfluously for the first 3 representations and parsimoniously for the 4th representation. When a variant has superfluous nucleotides on the left side, it is defined as not being left parsimonious as there is a need to left trim. The concept is symmetric for right parsimony and trimming. Parsimony applies to Indels too which we shall demonstrate in the left alignment section.
This figure shows multiple representations of a MNP. The left shows 4 possible representations differentiated by color. The right shows the corresponding representation in VCF. The last representation represents the parsimonious representation of the MNP.
Based on the definition of parsimony, it is easy to see that:
If a variant is non parsimonious, all its alleles must have length greater than 1.
However, the converse is not true.
Left alignment
Left aligning a variant means shifting the start position of that variant to the left till it is no longer possible to do so. It is a concept associated with insertion and deletion variants and describes specifically the nature of a position of a variant as opposed to its length. In order to further differentiate left alignment from simply left padding a variant, the definition is as follows:
A variant is left aligned if and only if it is no longer possible to shift its position to the left while keeping the length of all its alleles constant.
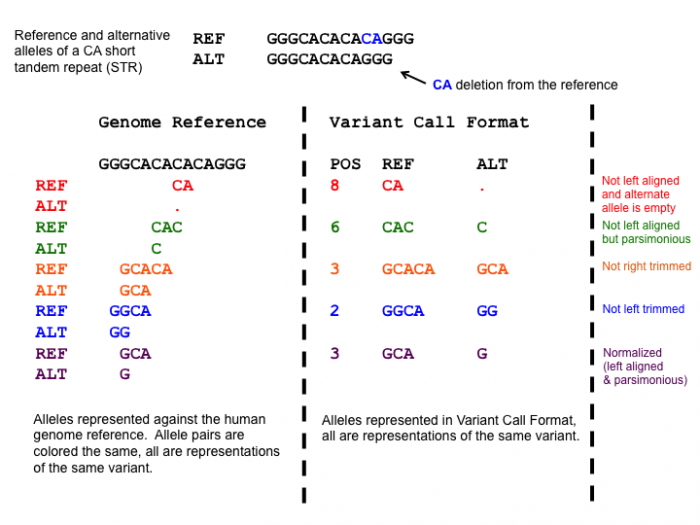
The following figure shows examples of an unnormalized short tandem repeat which is a special class of indels. The color of the text is synchronized to the variant that is described in the figure.
- The representation of variants in a VCF file requires that no alleles in the REF and ALT field are represented with an empty string (empty allele). The red indel has an illegal VCF representation.
- The green variant is not left aligned as you can prefix an A nucleotide on the left side of the variant's alleles and truncate the C on the right side of the variant's alleles. It is however parsimonious.
- The orange variant is left aligned but is not right parsimonious.
- The blue variant is left aligned but not left parsimonious.
- The maroon variant is left aligned and parsimonious.
This figure shows multiple representations of a CA tandem repeat. The left shows five possible representations differentiated by color. The right shows the corresponding representation in VCF. The last representation represents the left aligned and parsimonious representation of the STR.
When is a variant normalized?
A variant is normalized if and only if it is parsimonious and left aligned.
Lemma
In order to detect if a variant is normalized, we first prove the following lemma (1).
Each allele ends with the same nucleotide if and only if it is not left aligned or not right parsimonious.
For the => direction, we prove the contrapositive :
- If an indel is left aligned and right parsimonious then each allele does not end with the same type of nucleotide.
We first assume an indel is already left aligned and right parsimonious. Suppose all alleles have a length greater than 1, since the indel is right parsimonious; clearly, each allele do not end with the same type of nucleotide. Now, suppose that there exists an allele of length 1, and that all the alleles end with a particular nucleotide say 'A'. This is still considered right parsimonious as there are no superfluous nucleotides to remove without resulting in an empty allele. It is possible to extend all the alleles one position to the left by copying from a nucleotide on the reference genome, so now we have a superfluous nucleotide on the right side. Trimming off that nucleotide results in a new representation that shifts the indel to the left by one position while retaining the allele lengths of the original representation. This is left aligning the indel and thus there is a contradiction. Therefore each allele cannot end with the same type of nucleotide.
For the <= direction :
- If an indel is not left aligned or not right parsimonious then each allele ends with the same type of nucleotide.
Suppose a variant is not left aligned, then it must be possible to extend the alleles one nucleotide to the left and remove one nucleotide from the right to ensure that all the alleles remain the same length. Thus each allele must end with the same type of nucleotide for the removal of the rightmost nucleotide to be possible.
Suppose a variant is not right parsimonious, then by definition, the rightmost nucleotide is the same for all alleles which may be removed.
Corollary
A variant is normalized if and only if
1. it has no superfluous nucleotides on the left side and
2. each allele does not end with the same type of nucleotide.
Proof:
- A variant is normalized if and only if it is parsimonious and left aligned.
- ⇒ A variant is normalized if and only if it is left parsimonious and right parsimonious and left aligned. (break up parsimonious definition)
- ⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and right parsimonious and left aligned. (definition of left parsimony)
- ⇒ A variant is normalized if and only if it is has no superfluous nucleotides on the left side and each allele does not end with the same type of nucleotide. (lemma 1)
Uniqueness
It is important that the normalization results in a unique representation of the variant. Before we begin the proof, intuitively, accept that any representation of a variant can be transformed to another representation by adding nucleotides from the reference sequence to either ends of all the alleles at the same time or removing equivalent nucleotides from the ends of all the alleles at the same time.
Now suppose there are 2 normalized variants A and B. Suppose A is at a different position from B and B is to the right of A (without loss in generality), this is not possible because by the definition of a normalized variant, it is left aligned,
and if they were at different positions, that means B may be left aligned to A since they represent the same variants leading to a contradiction. So A and B must be at the same position.
Now, suppose that A and B are at the same position but are of different lengths where B is longer than A (without loss in generality), this is not possible as B is then not parsimonious, so B can be trimmed to the same length as A.
Thus A and B have to be at the same position and have the same length and variant normalization is unique.
Implementation
This is implemented in vt.
Algorithm for Normalization
So now that we know how to tell if a variant is normalized, we simply need to manipulate the variant till the rightmost ends of the alleles are not the same, and apply truncation to the superfluous nucleotides on the left side of the variant to obtain a normalized variant. The algorithm to normalize a biallelic or multiallelic variant is as follows:
Lines 1 to 8 performs the left alignment and ensures parsimonious representation on the right side. Lines 9 to 11 ensures parsimonious representation on the left side.
Comparisons
20 May 2014
The following table shows the number of variants normalized for an anonymous data set. This analysis was done on 20 May 2014.
| Dataset | bcftools | gatk | vt | comments |
|---|---|---|---|---|
| #normalized | - | 18794 | 18849 | bcftools's normalization is buggy, variants were truncated despite having differing prefix. |
| #normalized after bcftools | - | - | - | - |
| #normalized after gatk | - | 0 | 57 | 57 variants from GATK's normalization were left aligned by vt. 6 were biallelic and 51 were multiallelic. Note that 2 variants were changed by GATK but were not completely normalized. |
| #normalized after vt | - | 0 | 0 | no variants processed by vt were further normalized. |
Commands used are:
- bcftools norm -f ref.fa in.vcf -O z > out.vcf.gz
- java -jar GenomeAnalysisTK.jar -T LeftAlignAndTrimVariants --trimAlleles -R ref.fa --variant in.vcf.gz -o out.vcf.gz
- vt normalize -r ref.vcf.gz -o out.vcf.gz
Versions are:
- bcftools v0.2.0-rc8-5-g0e06231 (using htslib 0.2.0-rc8-6-gd49dfa6)
- GATK v3.1-1-g07a4bf8
- vt normalize v0.5
Issues have been communicated to bcftools and gatk developers on 20 May 2014.
22 May 2014
The following table shows the number of variants normalized for an anonymous data set. This analysis was done on 22 May 2014. bcftools was updated.
| Dataset | bcftools | gatk | vt | comments |
|---|---|---|---|---|
| #normalized | 18849 | 18794 | 18849 | bcftools's algorithm is the same as vt now |
| #normalized after bcftools | 0 | 0 | 0 | no variants processed by bcftools were further normalized. |
| #normalized after gatk | 57 | 0 | 57 | GATK's algorithm is documented to work only for biallelic simple indels. The 57 variants were either multiallelic or mixed variants (SNP adjacent to indel). 6 biallelic mixed variants that were not left aligned however turned out to be simple indels after left alignment. |
| #normalized after vt | 0 | 0 | 0 | no variants processed by vt were further normalized. |
Commands used are:
- bcftools norm -f ref.fa in.vcf -O z > out.vcf.gz
- java -jar GenomeAnalysisTK.jar -T LeftAlignAndTrimVariants --trimAlleles -R ref.fa --variant in.vcf.gz -o out.vcf.gz
- vt normalize -r ref.vcf.gz -o out.vcf.gz
Versions are:
- bcftools v0.2.0-rc8-5-g0e06231 (using htslib 0.2.0-rc8-6-gd49dfa6) [updated non release development version]
- GATK v3.1-1-g07a4bf8
- vt normalize v0.5
Here is an example where this normalization algorithm fails
We distinguish the concepts of normalization and decomposition/reconstruction of variants as follows:
Normalization involves reducing representations of a variant to a canonical representation. Normalization can be applied to biallelic variants or multiallelic variants. The problem of normalization is solvable and there exists a unique representation that is left aligned and parsimonious. Mathematical proof is published. [1]
Decomposition of variants involves the breaking down of a variant record into multiple records. It may be done vertically - as in multiallelics becoming biallelics or it can be done horizontally - a cluster of indels and SNPs represented as a complex variant being splitted up into several records. Horizontal decompositions in general do not have a unique solution. Similarly, reconstruction combines several variant records into a single record and can be done vertically and horizontally too. Vertical decomposition of a multiallelic variant to a set of biallelic records is a many to one function. Construction of a set of biallelic variants into a multiallelic record is not unique as you need to considered all possible permutations of the haplotypes containing your alleles.
If your example contains the decomposition or reconstruction of variants, then it is probable that you can find inconsistencies.
It is important to distinguish the difference between normalization and decomposition/reconstruction. The notion of normalization implies that a variant can be reduced to a standardized form. If you were to include decomposition and reconstruction in your notion of normalization, you are bound to find inconsistencies simply due to the inherent issues of identifiability.
When performing decomposition and construction, I think the following factors should be considered:
- Are your variants describing just a single individual or a population?
- Are the genotypes (if any) in your individual(s) phased?
Depending on the context, you will obtain different answers.
An example of inconsistent variant representation due to using vt normalize
Citation
Translations
A mandarin translation can be found here
Maintained by
This page is maintained by Adrian.