Difference between revisions of "Variant Normalization"
| Line 1: | Line 1: | ||
= Introduction = | = Introduction = | ||
| − | Variant representation in Variant Call Format is non unique. We describe a variant normalization here that is parsimonious and left aligned. | + | Variant representation in Variant Call Format is non-unique. We describe a variant normalization here that is parsimonious and left aligned. |
= Normalization = | = Normalization = | ||
Revision as of 15:54, 9 July 2013
Introduction
Variant representation in Variant Call Format is non-unique. We describe a variant normalization here that is parsimonious and left aligned.
Normalization
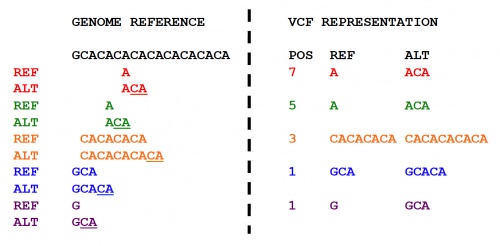
This figure shows multiple representations of a CA tandem repeat. The left shows five possible representations differentiated by color. The right shows the corresponding representation in VCF. The last representation represents the left aligned and parsimonious representation of the Indel.
Parsimony
This refers to the representation of a variant in as few nucleotides as possible. A representation with nucleotides that can be inferred from the reference sequence is not considered parsimonious. The blue variant representation in the figure is non parsimonious while the maroon representation is.
Left alignment
Left alignment is usually a concept associated with Indels. We define an indel to be left aligned when the variant can not be shifted to the left any further while ensuring that the indel represented is consistent and that no alleles are represented with an empty string (empty allele). The orange representation is not left aligned while the blue representation is.
How to observe that a variant is not left aligned or parsimonious on the right side?
If the ends of each allele is the same nucleotide, it is not left aligned or parsimonious on the right side.
Proof of left alignment
Suppose an indel is already left aligned. In order to shift the variant to the right, we have to be able to truncate the first leftmost nucleotide in each allele without any loss of information (i.e. we can reconstruct the original alleles from the right aligned version of the variant given the reference genome). In order to guarantee this, the first leftmost nucleotide in each allele should be the same type of nucleotide (in other words, the same as the reference nucleotide).
The truncation of the first leftmost nucleotide should not result in any empty allele. To achieve this, we need to first extend the rightmost end of each allele by the base observed on the reference sequence and then attempting to truncate the alleles simultaneously on the leftmost end. Thus, an indel is not left aligned if the rightmost nucleotide of each allele is represented by the same type of nucleotide. This procedure also ensures parsimony on the right side of the variants.
Algorithm for Normalization
The algorithm to normalize a variant; biallelic or multiallelic is as follows:
Lines 1 to 3 ensures parsimonious representation on the left side. Lines 4 to 9 performs the left alignment and ensures parsimonious representation on the right side. In the case of STRs, we might prefer the repeat units be retained.