SeqShop: Calling Your Own Genome, December 2014
Login instructions for seqshop-server
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
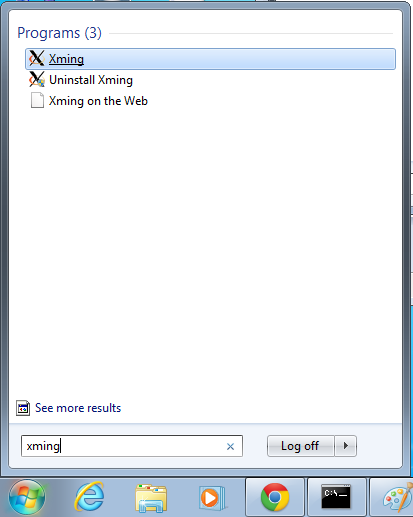
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
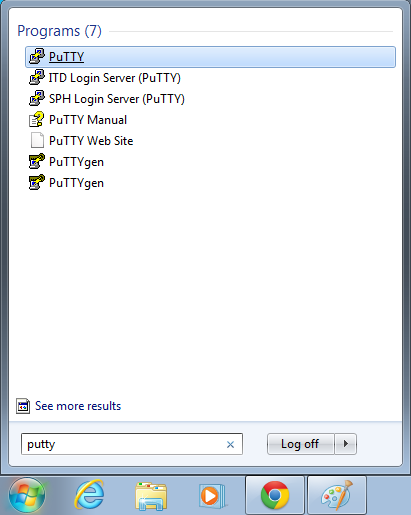
- Start->Enter "putty" in the search and select "PuTTY" from the program list
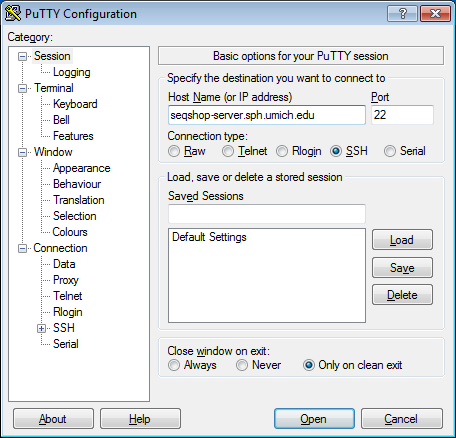
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
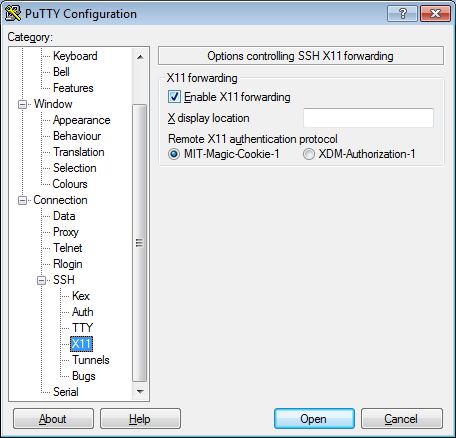
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup
The snpcall pipeline will run overnight, but you'll want to log out.
- How do I leave something running on the server even if I log out?
- One solution is screen!
- How do I use screen?
- Before running your command, you need to start screen:
screen
As it says, press Space or Return.
- It should now look basically the same as your normal command line.
- Scrolling problems when using screen?
- If you want to scroll and screen doesn't scroll like you normally would?
- Type Ctrl-a Esc and you should be able to scroll up with your mouse wheel
- Or at least that is what I do from my Linux machine - (sorry I'm typing this up/testing these commands from Linux and not windows, so can't test it out)
- Type Ctrl-a Esc and you should be able to scroll up with your mouse wheel
Set these values. Also, be sure to specify your sample name instead of SampleXX
export SAMPLE=SampleXX source /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
See the settings you just used:
cat /net/seqshop-server/home/mktrost/seqshop/setupSS.txt
Shows you:
export GC=/net/seqshop-server/home/mktrost/seqshop/gotcloudexport OUT=~/$SAMPLE/output
List of BAMs
The list of BAMs has already been created (just 1 BAM, your sample).
- But it is simply SAMPLE\tBAM_name, so easy to figure out
cat $SAMPLE/output/bam.list
SampleXX SampleXX/output/bams/SampleXX.recal.bam
- Relative path, so assumes running from your home directory (I prefer absolute paths, but for simplicity of the workshop, we just use relative path).
Configuring SnpCall
cat $SAMPLE/gotcloud.conf
You will see something like this:
# Cluster Settings BATCH_TYPE = BATCH_OPTS = OUT_DIR = Sample13/output # Align Settings MAP_TYPE = BWA_MEM BWA_THREADS = -t 24 FASTQ_LIST = fastq.list # SNP Call Settings UNIT_CHUNK = 20000000 # Chunk size of SNP calling : 20Mb VCF_EXTRACT = /net/seqshop-server/home/mktrost/seqshop/singleSample/snpOnly.vcf.gz MODEL_GLFSINGLE = TRUE MODEL_SKIP_DISCOVER = FALSE MODEL_AF_PRIOR = TRUE EXT_DIR = /net/seqshop-server/home/mktrost/seqshop/singleSample/ext EXT = $(EXT_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(EXT_DIR)/chrCHR.filtered.sites.vcf.gz
Running SnpCall
${GC}/gotcloud snpcall --conf $SAMPLE/gotcloud.conf --numjobs 6
- Only need the configuration & number of threads, rest is specified within the configuration.
Run 4 jobs on our mini-cluster
$GC/gotcloud snpcall --conf ~/personal/gotcloud.2x.conf --numjobs 4 --batchtype mosix --batchopts "-j10,11,12,13"
- --batchtype says to use mosix (our cluster system)
- --batchopts tells mosix the options to run with
- for mosix, -j10,11,12,13 says to run on nodes 10, 11, 12, & 13 - the names of the 4 nodes on our mini-cluster
Exome
To speed things up, I extracted only exome regions from 100 1000g low coverage BAMs.
Let's create a new bam info file with your BAM combined with those BAMs.
cp $ALIGN_OUT/bam.index $EXOME_OUT/bam.exome.index
Now add the exome BAMs to your new bam list:
cat $IN/exome/bam.exome.index >> $EXOME_OUT/bam.exome.index
Verify you have 101 lines in your list:
wc -l $EXOME_OUT/bam.exome.index
We are going to run on the cluster, so edit the first line of $EXOME_OUT/bam.exome.index to give the cluster path to your info file.
nedit $EXOME_OUT/bam.exome.index
Replace the /home on the first line with /net/seqshop-server
Locate your gotcloud.2x.conf (probably at: ~/personal/gotcloud.2x.conf) and open it in your favorite editor:
nedit ~/personal/gotcloud.2x.conf
Replace all occurrances of
/home with /net/seqshop-server
This is so you can run on the mini-cluster we have and can run more jobs at once
Update OUT_DIR & BAM_INDEX to:
OUT_DIR = $(IN_DIR)/output.exome BAM_INDEX = $(OUT_DIR)/bam.exome.index
Update your gotcloud configuration file to indicate exomes:
# Specify the path to the regions we want to call UNIFORM_TARGET_BED = $(REF_DIR)/20130108.exome.targets.nochr.bed # We do not want any off target bases OFFSET_OFF_TARGET = 0 WRITE_TARGET_LOCI = TRUE TARGET_DIR = target
Remove CHRS = 20
Since it would take a while to run all 101 samples, I already ran the first step for the 100 1000G samples. We will "trick" GotCloud into thinking you already ran them by copying them into your output directory.
cp -r $IN/exome/glfs $EXOME_OUT/.
Run 4 jobs on our mini-cluster
$GC/gotcloud snpcall --conf ~/personal/gotcloud.2x.conf --numjobs 4 --batchtype mosix --batchopts "-j10,11,12,13"
- --batchtype says to use mosix (our cluster system)
- --batchopts tells mosix the options to run with
- for mosix, -j10,11,12,13 says to run on nodes 10, 11, 12, & 13 - the names of the 4 nodes on our mini-cluster
Log Out
- Want to log out and leave your job running?
In the screen window, type:
Ctrl-a d
(Hold down Ctrl and type 'a', let go of both and type 'd')
- This will "detach" from your screen session while your alignment continues to run.
If you have not detached from screen:
Ctrl-a d
exit PuTTY
Logging Back in to Check Jobs
- How do you log back into screen tomorrow?
screen -r
This will resume an already running screen.