SeqShop: Calling Your Own Genome, June 2014
Login to the seqshop-server Linux Machine
This section will appear redundantly in each session. If you are already logged in or know how to log in to the server, please skip this section
- Login to the windows machine
- The username/password for the Windows machine should be written on the right-hand monitor
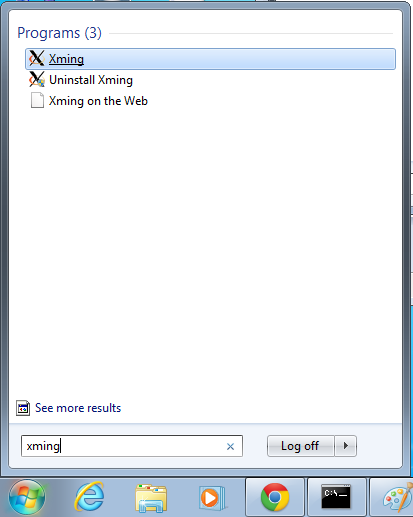
- Start->Enter "Xming" in the search and select "Xming" from the program list
- Nothing will happen, but Xming was started.
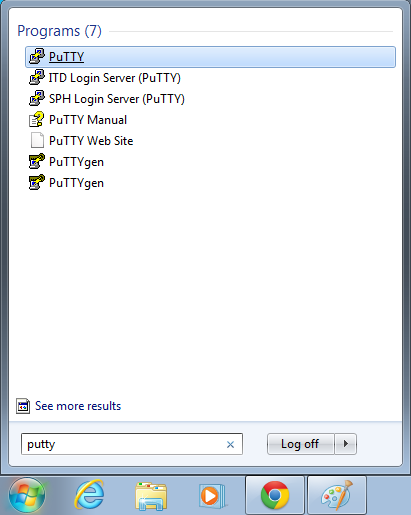
- Start->Enter "putty" in the search and select "PuTTY" from the program list
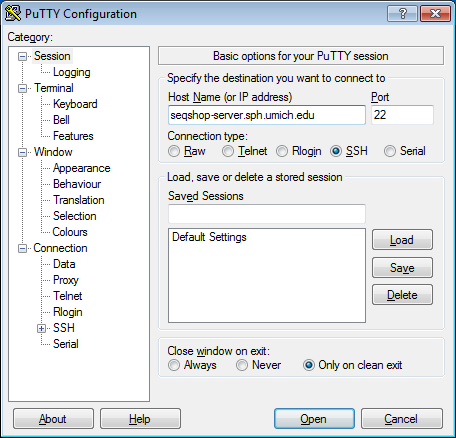
- Host Name:
seqshop-server.sph.umich.edu - Setup to allow you to open external windows:
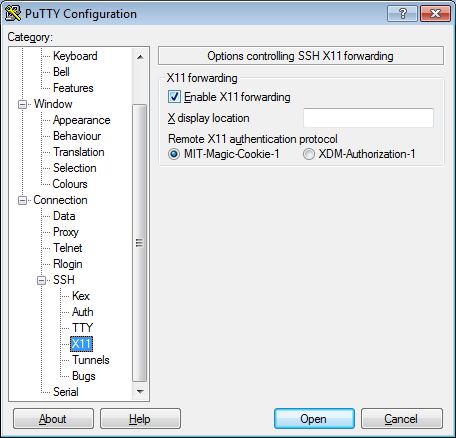
- In the left pannel: Connection->SSH->X11
- Add a check mark in the box next to
Enable X11 forwarding - Click
Open - If it prompts about a key, click
OK
You should now be logged into a terminal on the seqshop-server and be able to access the test files.
- If you need another terminal, repeat from step 3.
Login to the seqshop Machine
So you can each run multiple jobs at once, we will have you run on 4 different machines within our seqshop setup.
- You can only access these machines after logging onto seqshop-server
3 users logon to:
ssh -X seqshop1
3 users logon to:
ssh -X seqshop2
2 users logon to:
ssh -X seqshop3
2 users logon to:
ssh -X seqshop4
Setup
Set these values. If you used a different path for any of these, please update here. Also, be sure to specify your sample name instead of Sample_XXXXX
source /home/mktrost/seqshop-server/setup.txt export SAMPLE=Sample_XXXXX
OUT needs to point to where your alignment output went, so if your output is not ~/personal/output, please set OUT appropriately:
export OUT=~/personal/YOUR_OUTPUT_DIR
Verify that this does not give an error:
ls $OUT/bams/${SAMPLE}.recal.bam
GotCloud Configuration Updates
To speed things up, we will only run on certain regions
- We need to update the GotCloud Configuration to do this.
Locate your gotcloud.2x.conf (probably at: ~/personal/gotcloud.2x.conf) and open it in your favorite editor:
nedit ~/personal/gotcloud.2x.conf
Add
# Specify the path to the regions we want to call UNIFORM_TARGET_BED = $(REF_DIR)/20130108.exome.targets.nochr.bed # We do not want any off target bases OFFSET_OFF_TARGET = 0 WRITE_TARGET_LOCI = TRUE TARGET_DIR = target
Run
Set CHR = 1 2
cp -r /home/mktrost/seqshop/inputs/glfs personal/output/.
cp personal/output/bam.index personal/output/bam.exome.index
cat /home/mktrost/seqshop/inputs/20130502.gotcloud.low_coverage.100.index >> personal/output/bam.exome.index
wc -l personal/output/bam.exome.index
cp /home/mktrost/seqshop/inputs/gotcloud.exome.conf personal/.
Update the conf ...your home.
/home/mktrost/seqshop/gotcloud/gotcloud snpcall --conf personal/gotcloud.exome.conf