Difference between revisions of "Understanding vcf-summary output"
From Genome Analysis Wiki
Jump to navigationJump to search(No difference)
|
Revision as of 22:46, 16 June 2014
What is vcf-summary?
vcf-summary is a utility included in GotCloud that helps evaluate the quality of SNP calls. Because GotCloud will automatically run vcf-summary, detailed instructions on the usage of the program is not currently documented.
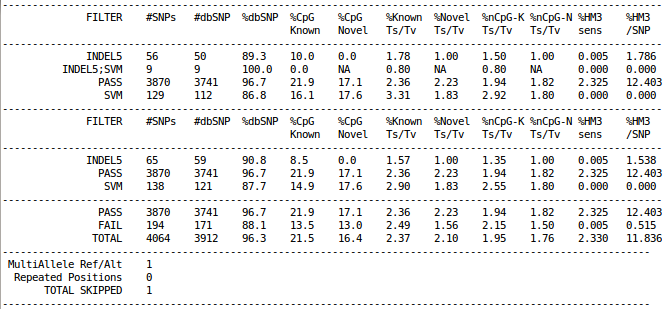
Example output from vcf-summary
If OUT is an environment variable, you may see output file from GotCloud similar to the following example.
cat ${OUT}/vcfs/chr22/chr22.filtered.sites.vcf.summary
The example above is obtained from the results of GotCloud within a very small (1Mb) region in chr22 across ~60 1000 Genomes samples.
Rows of vcf-summary output has three sections
As shown in the example figure above, typical vcf-summary output primarily consists of the following three sections.
- In the first part, each SNP is counted only once, grouped by the contents of FILTER column.
- In the second part, each SNP may be counted multiple times, if the SNP failed multiple filters (e.g. INDEL5 filter and SVM filter).
- In the last part, each SNP is counted only once, grouped by SNPs with "PASS" in the FILTER column versus everything else.
In addition, multi-allelic or duplicated SNPs are counted separately at the very bottom.