Difference between revisions of "SeqShop: Sequence Mapping and Assembly Practical, June 2014"
| Line 1: | Line 1: | ||
| − | == Step 0: Login to the machine == | + | == Step 0: Login to the machine & setup environment== |
# Login to the windows machine | # Login to the windows machine | ||
| Line 9: | Line 9: | ||
#* Server name: seqshop-server.sph.umich.edu | #* Server name: seqshop-server.sph.umich.edu | ||
#* Enter your provided username & password | #* Enter your provided username & password | ||
| + | # To simplify commands/typing, we will setup an environment variable to point to the GotCloud directory. | ||
| + | export GC=/home/mktrost/seqshop/ | ||
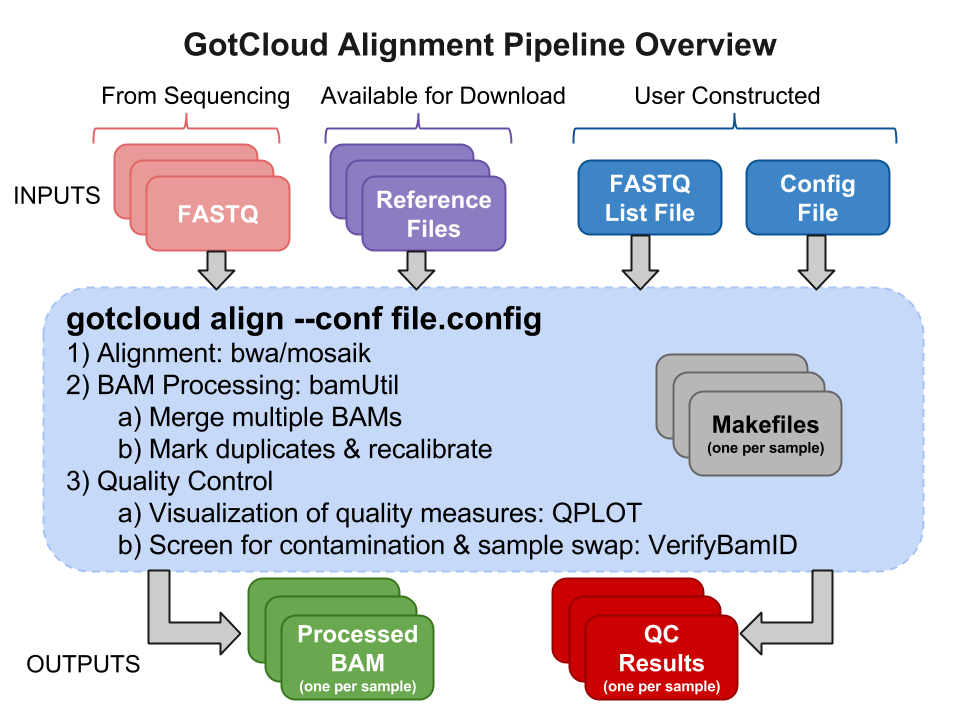
== GotCloud Alignment Pipeline== | == GotCloud Alignment Pipeline== | ||
| − | |||
| − | |||
| − | + | [[File:AlignDiagram.png]] | |
| − | + | ||
=== Input Files === | === Input Files === | ||
| − | + | ==== Sequence Data Files : FASTQs ==== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==== Sequence | ||
The FASTQ files are provided to you by those who did the sequencing. | The FASTQ files are provided to you by those who did the sequencing. | ||
For this tutorial, we will use FASTQs for 6 1000Genome samples | For this tutorial, we will use FASTQs for 6 1000Genome samples | ||
ls ${GC}/inputs/fastq/ | ls ${GC}/inputs/fastq/ | ||
| − | There are 51 fastq files. | + | There are 51 fastq files: combination of single-end & paired-end. |
| + | * Single-end: HG00641.chr7.CFTR.SRR069531.fastq | ||
| + | * Paired-end: HG00641.chr7.CFTR.SRR069531'''_1'''.fastq & HG00641.chr7.CFTR.SRR069531'''_2'''.fastq | ||
| + | |||
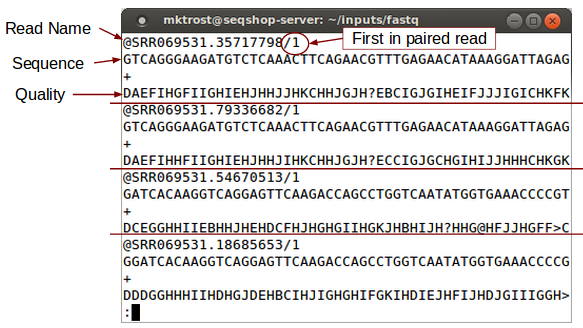
| + | Look at FASTQ: | ||
| + | less -S ${GC}/inputs/fastq/HG00641.chr7.CFTR.SRR069531.fastq | ||
| + | <code>less</code> is a Linux command that allows you to look at a file. | ||
| + | *<code>-S</code> option prevents line wrap. | ||
| + | * Use the arrow (up/down/left/right) keys to scroll through the file. | ||
| + | * Use <code>zless</code> if the file is compressed. | ||
| + | |||
| + | [[File:FASTQ.png]] | ||
| + | |||
| + | |||
| + | |||
| + | ==== Reference Files ==== | ||
| + | Reference files can be downloaded with GotCloud or from other sources. | ||
| + | |||
| + | ls ${GC}/reference/chr7 | ||
| + | |||
| + | |||
| + | ==== GotCloud FASTQ Index File ==== | ||
| + | This file is created by you and directs GotCloud to your FASTQ files and provides additional information for them. | ||
| + | |||
| + | Tab delimited file that starts with a header line. The columns may be in any order. | ||
| + | |||
| + | Following the header line, there is one line per single-end read and one line per paired-end read (only 1 line per pair). | ||
| + | |||
| + | Required Column Names: | ||
| + | * MERGE_NAME - base name for the resulting BAM file for the sample (used to group multiple fastqs or fastq pairs into a single BAM) | ||
| + | * FASTQ1 - name of the fastq or the first in the pair if paired-end. (Only 1 line per pair) | ||
| + | |||
| + | Optional Column Names: | ||
| + | * FASTQ2 - name of the 2nd fastq in paired-end reads. Specify '.' if the column exists, but this line is single-ended. | ||
| + | * RGID - Read Group ID for this entry | ||
| + | * SAMPLE - Sample Name for this entry | ||
| + | * LIBRARY - Library for this entry | ||
| + | * CENTER - Center Name for this entry | ||
| + | * PLATFORM - Platform for this entry | ||
| + | |||
| + | The RGID, SAMPLE, LIBRARY, CENTER, and PLATFORM are used to populate the Read Group information for this entry. These fields are optional. Either leave the column header out of the file or specify '.' if the column header exists, but the data is N/A. As long as the RGID field is specified non-N/A fields are added to the BAM file. | ||
| + | |||
| + | MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM | ||
| + | Sample1 fastq/S1/F1_R1.fastq.gz fastq/S1/F1_R2.fastq.gz RGID1 SampleID1 Lib1 UM ILLUMINA | ||
| + | Sample1 fastq/S1/F2_R1.fastq.gz fastq/S1/F2_R2.fastq.gz RGID1a SampleID1 Lib1 UM ILLUMINA | ||
| + | Sample2 fastq/S2/F1_R1.fastq.gz fastq/S2/F1_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA | ||
| + | Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA | ||
| + | |||
| + | The <code>--fastq</code>/<code>FASTQ</code> setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files. | ||
| + | |||
| + | What is a Read Group? | ||
| + | * Groups reads together | ||
| + | * Used for recalibration | ||
| + | ** Each sequencing run should get a different ReadGroup | ||
| + | |||
| + | This file is specified either via the command line parameter <code>--index_file</code> or via the configuration file setting <code>INDEX_FILE</code>. | ||
| + | |||
| + | The command-line setting takes precedence over the configuration file setting. | ||
| + | |||
| + | |||
| + | ==== GotCloud Configuration File ==== | ||
| + | This file is created by you to configure GotCloud for your data. | ||
Revision as of 22:33, 9 June 2014
Step 0: Login to the machine & setup environment
- Login to the windows machine
- The username/password for the Windows machine should be written on it
- Open putty
- Start->.....
- In putty, login to seqshop-server.sph.umich.edu
- Server name: seqshop-server.sph.umich.edu
- Enter your provided username & password
- To simplify commands/typing, we will setup an environment variable to point to the GotCloud directory.
export GC=/home/mktrost/seqshop/
GotCloud Alignment Pipeline
Input Files
Sequence Data Files : FASTQs
The FASTQ files are provided to you by those who did the sequencing.
For this tutorial, we will use FASTQs for 6 1000Genome samples
ls ${GC}/inputs/fastq/
There are 51 fastq files: combination of single-end & paired-end.
- Single-end: HG00641.chr7.CFTR.SRR069531.fastq
- Paired-end: HG00641.chr7.CFTR.SRR069531_1.fastq & HG00641.chr7.CFTR.SRR069531_2.fastq
Look at FASTQ:
less -S ${GC}/inputs/fastq/HG00641.chr7.CFTR.SRR069531.fastq
less is a Linux command that allows you to look at a file.
-Soption prevents line wrap.- Use the arrow (up/down/left/right) keys to scroll through the file.
- Use
zlessif the file is compressed.
Reference Files
Reference files can be downloaded with GotCloud or from other sources.
ls ${GC}/reference/chr7
GotCloud FASTQ Index File
This file is created by you and directs GotCloud to your FASTQ files and provides additional information for them.
Tab delimited file that starts with a header line. The columns may be in any order.
Following the header line, there is one line per single-end read and one line per paired-end read (only 1 line per pair).
Required Column Names:
- MERGE_NAME - base name for the resulting BAM file for the sample (used to group multiple fastqs or fastq pairs into a single BAM)
- FASTQ1 - name of the fastq or the first in the pair if paired-end. (Only 1 line per pair)
Optional Column Names:
- FASTQ2 - name of the 2nd fastq in paired-end reads. Specify '.' if the column exists, but this line is single-ended.
- RGID - Read Group ID for this entry
- SAMPLE - Sample Name for this entry
- LIBRARY - Library for this entry
- CENTER - Center Name for this entry
- PLATFORM - Platform for this entry
The RGID, SAMPLE, LIBRARY, CENTER, and PLATFORM are used to populate the Read Group information for this entry. These fields are optional. Either leave the column header out of the file or specify '.' if the column header exists, but the data is N/A. As long as the RGID field is specified non-N/A fields are added to the BAM file.
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM Sample1 fastq/S1/F1_R1.fastq.gz fastq/S1/F1_R2.fastq.gz RGID1 SampleID1 Lib1 UM ILLUMINA Sample1 fastq/S1/F2_R1.fastq.gz fastq/S1/F2_R2.fastq.gz RGID1a SampleID1 Lib1 UM ILLUMINA Sample2 fastq/S2/F1_R1.fastq.gz fastq/S2/F1_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA
The --fastq/FASTQ setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files.
What is a Read Group?
- Groups reads together
- Used for recalibration
- Each sequencing run should get a different ReadGroup
This file is specified either via the command line parameter --index_file or via the configuration file setting INDEX_FILE.
The command-line setting takes precedence over the configuration file setting.
GotCloud Configuration File
This file is created by you to configure GotCloud for your data.