Difference between revisions of "SeqShop: Sequence Mapping and Assembly Practical, June 2014"
| Line 45: | Line 45: | ||
==== GotCloud FASTQ Index File ==== | ==== GotCloud FASTQ Index File ==== | ||
| − | This file is created by you and directs GotCloud to your FASTQ files | + | This file is created by you and directs GotCloud to your FASTQ files, providing additional information for them. |
| − | + | * tab delimited | |
| + | * columns may be in any order | ||
| + | * starts with a header line | ||
| + | * one line per single-end read | ||
| + | * one line per paired-end read (only 1 line per pair). | ||
| − | + | Required Columns | |
| + | {|class="wikitable" cellpadding=5 | ||
| + | ! Column Name !! Description !! Recommended Value | ||
| + | |- | ||
| + | | MERGE_NAME || | ||
| + | * Base name for the resulting BAM file for the sample | ||
| + | * Used to group multiple fastqs or fastq pairs into a single BAM | ||
| + | | Sample Name | ||
| + | |- | ||
| + | | FASTQ1 || | ||
| + | * Name of the fastq or the first in the pair if paired-end. (Only 1 line per pair) | ||
| + | | path/fastq1 | ||
| + | |- | ||
| + | | FASTQ2 || | ||
| + | *Name of the 2nd fastq in paired-end reads. | ||
| + | *Column is not required if all fastqs are single-end | ||
| + | *'.' if the column is used, but this line is single-ended. | ||
| + | | path/fastq2 | ||
| + | |} | ||
| − | |||
| − | |||
| − | |||
| − | + | The following columns are optional and used to populate the Read Group Information in the BAM file. | |
| − | * | + | * RGID field is required if using any of these fields, the others are optional. |
| − | + | ||
| − | + | What is a Read Group? | |
| − | + | * Groups reads together | |
| − | + | * Used for recalibration | |
| − | + | ** Each sequencing run should get a different ReadGroup | |
| + | |||
| + | If you do not want the field for: | ||
| + | * any fastq, leave the column out of the header line | ||
| + | * a single line, use a '.' | ||
| + | |||
| + | {|class="wikitable" cellpadding=5 | ||
| + | |- | ||
| + | ! Column Name !! Description !! Recommended Value | ||
| + | |- | ||
| + | | RGID || Read Group ID || Run ID | ||
| + | |- | ||
| + | | SAMPLE || Sample Name || Sample Name | ||
| + | |- | ||
| + | | LIBRARY || Library | ||
| + | * separate FASTQs for a sample that were prepped separately | ||
| + | | if you don't know or it is all the same, use Sample Name | ||
| + | |- | ||
| + | | CENTER || Center Name || Name of the sequencing center producing the FASTQ | ||
| + | |- | ||
| + | | PLATFORM || Platform || CAPILLARY, LS454, ILLUMINA, | ||
| + | SOLID, HELICOS, IONTORRENT, or PACBIO | ||
| + | |} | ||
| + | |||
| − | |||
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM | MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM | ||
| Line 73: | Line 114: | ||
The <code>--fastq</code>/<code>FASTQ</code> setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files. | The <code>--fastq</code>/<code>FASTQ</code> setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files. | ||
| − | |||
| − | |||
| − | |||
| − | |||
This file is specified either via the command line parameter <code>--index_file</code> or via the configuration file setting <code>INDEX_FILE</code>. | This file is specified either via the command line parameter <code>--index_file</code> or via the configuration file setting <code>INDEX_FILE</code>. | ||
| − | The command-line setting takes precedence over the configuration file setting. | + | The command-line setting takes precedence over the configuration file setting. |
| − | |||
==== GotCloud Configuration File ==== | ==== GotCloud Configuration File ==== | ||
This file is created by you to configure GotCloud for your data. | This file is created by you to configure GotCloud for your data. | ||
Revision as of 16:37, 10 June 2014
Step 0: Login to the machine & setup environment
- Login to the windows machine
- The username/password for the Windows machine should be written on it
- Open putty
- Start->.....
- In putty, login to seqshop-server.sph.umich.edu
- Server name: seqshop-server.sph.umich.edu
- Enter your provided username & password
- To simplify commands/typing, we will setup an environment variable to point to the GotCloud directory.
export GC=/home/mktrost/seqshop/
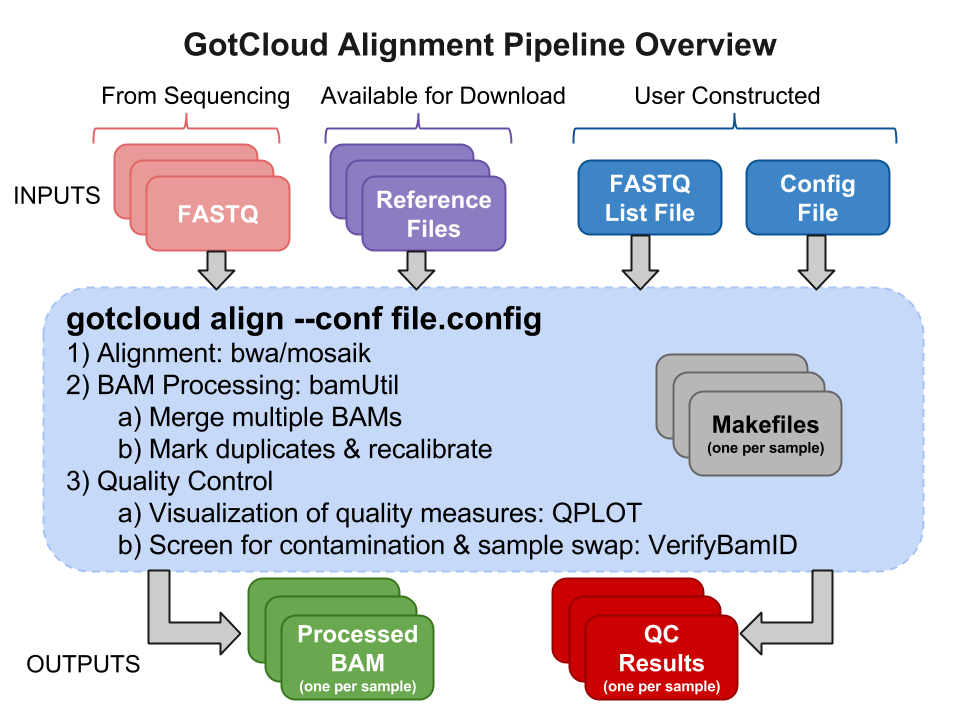
GotCloud Alignment Pipeline
Input Files
Sequence Data Files : FASTQs
The FASTQ files are provided to you by those who did the sequencing.
For this tutorial, we will use FASTQs for 6 1000Genome samples
ls ${GC}/inputs/fastq/
There are 51 fastq files: combination of single-end & paired-end.
- Single-end: HG00641.chr7.CFTR.SRR069531.fastq
- Paired-end: HG00641.chr7.CFTR.SRR069531_1.fastq & HG00641.chr7.CFTR.SRR069531_2.fastq
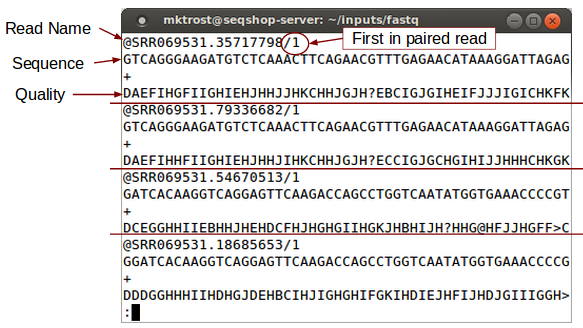
Look at FASTQ:
less -S ${GC}/inputs/fastq/HG00641.chr7.CFTR.SRR069531.fastq
less is a Linux command that allows you to look at a file.
-Soption prevents line wrap.- Use the arrow (up/down/left/right) keys to scroll through the file.
- Use
zlessif the file is compressed.
Reference Files
Reference files can be downloaded with GotCloud or from other sources.
ls ${GC}/reference/chr7
GotCloud FASTQ Index File
This file is created by you and directs GotCloud to your FASTQ files, providing additional information for them.
- tab delimited
- columns may be in any order
- starts with a header line
- one line per single-end read
- one line per paired-end read (only 1 line per pair).
Required Columns
| Column Name | Description | Recommended Value |
|---|---|---|
| MERGE_NAME |
|
Sample Name |
| FASTQ1 |
|
path/fastq1 |
| FASTQ2 |
|
path/fastq2 |
The following columns are optional and used to populate the Read Group Information in the BAM file.
- RGID field is required if using any of these fields, the others are optional.
What is a Read Group?
- Groups reads together
- Used for recalibration
- Each sequencing run should get a different ReadGroup
If you do not want the field for:
- any fastq, leave the column out of the header line
- a single line, use a '.'
| Column Name | Description | Recommended Value |
|---|---|---|
| RGID | Read Group ID | Run ID |
| SAMPLE | Sample Name | Sample Name |
| LIBRARY | Library
|
if you don't know or it is all the same, use Sample Name |
| CENTER | Center Name | Name of the sequencing center producing the FASTQ |
| PLATFORM | Platform | CAPILLARY, LS454, ILLUMINA,
SOLID, HELICOS, IONTORRENT, or PACBIO |
MERGE_NAME FASTQ1 FASTQ2 RGID SAMPLE LIBRARY CENTER PLATFORM Sample1 fastq/S1/F1_R1.fastq.gz fastq/S1/F1_R2.fastq.gz RGID1 SampleID1 Lib1 UM ILLUMINA Sample1 fastq/S1/F2_R1.fastq.gz fastq/S1/F2_R2.fastq.gz RGID1a SampleID1 Lib1 UM ILLUMINA Sample2 fastq/S2/F1_R1.fastq.gz fastq/S2/F1_R2.fastq.gz RGID2 SampleID2 Lib2 UM ILLUMINA Sample2 fastq/S2/F2.fastq.gz . RGID2 SampleID2 Lib2 UM ILLUMINA
The --fastq/FASTQ setting can be used to specify a prefix to the FASTQ1/FASTQ2 file paths that should be applied before using the files.
This file is specified either via the command line parameter --index_file or via the configuration file setting INDEX_FILE.
The command-line setting takes precedence over the configuration file setting.
GotCloud Configuration File
This file is created by you to configure GotCloud for your data.