GotCloud: Variant Calling Pipeline
Back to parent: GotCloud
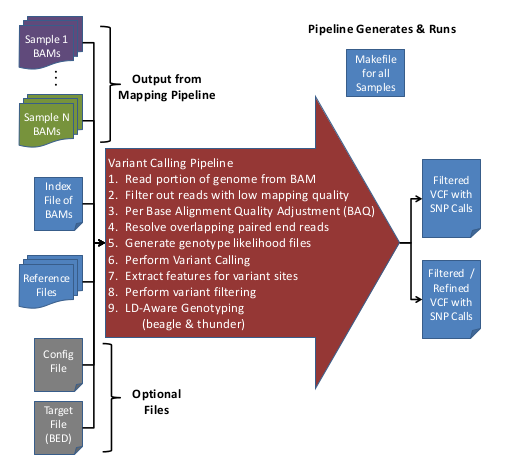
The Variant Calling Pipeline (previously called 'UMAKE') makes genotype calls from recalibrated BAM files. These genotype calls are output into VCF (Variant Call Format) files.
Running the GotCloud Variant Calling Pipeline
The variant calling pipeline (umake) is run using gotcloud snpcall and gotcloud ldrefine.
Running the Automatic Test
The automatic test runs the variant calling pipeline on a small test set and checks the results against expected results validating that GotCloud is installed correctly.
- Run
snpcallpipeline test:
gotcloud snpcall --test OUTPUT_DIR
- Where OUTPUT_DIR is the directory where you want to store the test results
- If you see
Successfully ran the test case, congratulations!, then you are ready to run snpcall on your own samples.
- Run
ldrefinepipeline test:
gotcloud ldrefine --test OUTPUT_DIR
- Where
OUTPUT_DIRis the directory where you want to store the test results - If you see
Successfully ran the test case, congratulations!, then you are ready to run ldrefine on your own samples.
- Where
Overview of Variant Calling Pipeline Steps
Here is an overview of the Variant Calling Pipeline:
For more information on the filters applied during the Variant Calling Pipeline, see, GotCloud: Filters.
Input Data
- Aligned/Processed/Recalibrated BAM files
- BAM list file containing Sample IDs & BAM file names
- Reference files
- (Optional) Configuration file to override default options
BAM Files
The BAM files need to be duplicate-marked and base-quality recalibrated in order to obtain high quality SNP calls. Generating these BAM files from original FASTQs is automatically done as part of the Alignment Pipeline of GotCloud.
BAM List File
- Automatically created when running the GotCloud Alignment Pipeline
- Each line of the BAM list file represents a single individual
Columns:
- sample id
- comma separated population labels (optional column)
- BAM File 1 (preferable to have full paths to BAM files)
- BAM File 2 (if more than 1 BAM per sample)
- ...
- # BAM File N (if more than 1 BAM per sample)
[SAMPLE_ID] [COMMA SEPARATED POPULATION LABELS] [BAM_FILE1] [BAM_FILE2] ...
or
[SAMPLE_ID] [BAM_FILE1] [BAM_FILE2] ...
- Notes:
- tab delimited
- multiple BAMs per individual may be provided, but should all be on the same line of the list file
- population label is optional - it will default to
ALL- only used by Thunder (part of ldrefine pipeline)
- if all samples are from the same population, population label can be skipped or you can just specify
ALLfor the population label for each sample.
The path to the BAM List file is defaulted to the outputDirectory/bam.list. It can be overridden by setting --bamlist, --bam_list, or --list on the command-line or by setting BAM_LIST in your configuration file to the path to the BAM List File. See Required Options for more information.
Reference Files
See GotCloud: Genetic Reference and Resource Files for detailed information about the multiple required reference files for the variant calling pipeline, including:
- How to obtain default references
- Configuration keys & default values
- How to generate your own references
- How to point GotCloud to your reference files
Required Reference File Types:
Configuration File
The GotCloud configuration file contains the run-time options, including software binaries and command line arguments. A default configuration file is automatically loaded. Users may specify their own configuration file specifying just the values different than the defaults. The configuration file is not required if there are no values to override.
- Default GotCloud configuration file is
gotcloud/bin/gotcloudDefaults.conf - Comments begin with a
# - Format:
KEY = value- where
KEYis the item being set andvalueis its new value
- where
- Some settings can be defined both in the configuration file and on the GotCloud command-line
- command-line options take priority over configuration file settings
- A KEY can be used in another KEY's value by specifying $(KEY)
- Example:
KEY1 = value1KEY2 = $(KEY1)/value2
- When
KEY2is used, it will be equal to:value1/value2
- Example:
Output Directory
- The output directory is required for running GotCloud, so GotCloud knows where to write its output
| Configuration Key | Command-line Flag | Value Description | ||
|---|---|---|---|---|
| OUT_DIR | --outdir | output directory | ||
Reference/Resource Files
- See GotCloud: Genetic Reference and Resource Files for reference/resource file configuration settings
Cluster Configuration
Regardless of the type of cluster system used, GotCloud will wait for each job to complete after launching it.
- For any BATCH_TYPEs that run in batch mode, GotCloud generates a script that will wait until the step is complete before returning
- In a sense, it "fakes" interactive mode for all batch types since it will not proceed until a command is finished
- If you are at UM and are using flux, you can specify either
fluxorpbs
| Configuration Key | Command-line Flag | Value Description | ||
|---|---|---|---|---|
| BATCH_TYPE | --batchtype | type of cluster system | ||
| Valid Values | Command to Launch | Command to Check for Completion | ||
mosix |
mosbatch -E/tmp |
N/A - interactive type | ||
sge |
qsub |
qstat -u $USER
| ||
sgei |
qrsh -now n |
N/A - interactive type | ||
pbs |
qsub |
qstat -u $USER
| ||
slurm |
sbatch |
squeue -u $USER
| ||
slurmi |
|
N/A - interactive type | ||
local |
N/A - local command | N/A - interactive type | ||
| BATCH_OPTS | --batchopts | options to pass to your cluster type, example:
-j36,37,38,39,40,41,45,46,47,48,49 | ||
See Variant Calling Command-line Options/Configuration Settings for more information on Configuration options.
Example Configuration File
Example configuration file where reference files happen to be stored in /path/reference, and bam index file in path/freeze5
CHRS = 20 22 BAM_LIST = /path/freeze5.bam.list OUT_DIR = /path/freeze5/output REF_DIR = /path/reference/ REF = $(REF_DIR)/hs37d5.fa INDEL_PREFIX = $(REF_DIR)/1kg.pilot_release.merged.indels.sites.hg19 HM3_VCF = $(REF_DIR)/hapmap3_r3_b37.sites.vcf.gz DBSNP_VCF = $(REF_DIR)/dbsnp_135.b37.sites.vcf.gz
Variant Calling Command-line Options/Configuration Settings
Required Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --outdir path | OUT_DIR | output directory | |
| --list/--bam_list/--bamlist file | BAM_LIST | path to the BAM List File | $(OUT_DIR)/bam.list |
| --numjobs # | number of jobs to run in parallel | 0 (generate Makefile of steps, but do not run) |
Common Options
| Common Options | |||
|---|---|---|---|
| Command-line Flag | Configuration Key | Value Description | Default Value |
| --conf file | configuration file to use | ||
Cluster Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --batchtype type | BATCH_TYPE | name of cluster type | local |
| --batchopts opts | BATCH_OPTS | options to pass to the cluster command | |
| --copyglf path | COPY_GLF | path to copy glfs to before processing them (path local to remote nodes, maybe in /tmp) |
Test/Debug Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --help | print help information | ||
| --test path | run the snpcall/ldrefine test and write output to the specified path | ||
| --verbose | Add additional messages when reading configuration |
Reference/Resource Files
- See GotCloud: Genetic Reference and Resource Files for reference/resource file configuration settings
Analysis Region Options
See Targeted/Exome Sequencing Settings for more information on specifying exome/targetted regions and other settings.
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --chrs # # | CHRS | pace separated list of chromosomes to process | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X |
| --region #:#-# | call region - skip regions of chromosome outside of specified region
format (-end is optional): chr:start-end |
||
| UNIT_CHUNK | chunk size of SNP calling (GotCloud breaks up each chromosome into regions of this size) | 5000000 | |
| LD_NSNPS | chunk size (number of SNPs) of genotype refinement | 10000 | |
| LD_OVERLAP | overlapping # of SNPs between chunks for genotype refinement | 1000 |
Chromosome X/Y Calling
For proper Chromosome X/Y calling, it is recommended to specify a PED file with sex information:
| Configuration Key | Value Description |
|---|---|
| PED_INDEX | ped file containing sampleID (2nd column) and sex (5th column) |
Format of PED file:
familyID sampleID fatherID motherID sex
- Only
sampleIDandsexare used
Targeted/Exome Sequencing Settings
If you are running Targeted/Exome Sequencing, the user should specify:
| Configuration Key | Value Description | |
|---|---|---|
| UNIFORM_TARGET_BED | Bed file of targeted regions (same bed for all samples) | |
| MULTIPLE_TARGET_MAP | Filename of file mapping: sample id -> bed file of targeted regions
Each line of the file contains: [SM_ID] [TARGET_BED] | |
| OFFSET_OFF_TARGET | Number of bases by which to extend the target region
(default is 0, do not extend the target region) | |
| SAMTOOLS_VIEW_TARGET_ONLY | true: speeds up processing by excluding off-target regions initially when performing samtools view
false (default): off-target regions are not excluded when performing samtools view, but are excluded at a later step Warning: You may not want to set this to true due to it may:
| |
| WGS_SVM | whether or not to run SVM on the whole genome rather than by chromosome (default is by chromosome). Set to TRUE if you are running with a small number of target regions. |
Path Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --makebasename name | MAKE_BASE_NAME | basename of the Makefile generated by GotCloud | umake |
| --bamprefix prefix | BAM_PREFIX | path to prepend to relative BAM file paths in the BAM list | |
| --refprefix prefix | REF_PREFIX | path to prepend to relative reference/resource file paths | |
| --baseprefix prefix | BASE_PREFIX | path to prepend to relative paths for the BAM list file, PED_INDEX, BAM (if BAM_PREFIX isn't specified), reference/resource files (if REF_PREFIX isn't specified) | |
| --refdir path | REF_DIR | value to use for REF_DIR key | $(GOTCLOUD_ROOT)/gotcloud.ref |
| --gotcloudroot path | GOTCLOUD_ROOT | specify to use a different directory for finding GotCloud bins/scripts | based on the location of the gotcloud/umake.pl script |
Validation Adjustment Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --maxlocaljobs # | maximum # of jobs that can run if batchtype is local (to prevent accidentally starting jobs locally that were meant to be on a cluster) | 10 | |
| --ignoresmcheck | IGNORE_SM_CHECK | disable the validation that the Sample name in the BAM file matches the one in the BAM list file |
Miscellaneous Options
| Command-line Flag | Configuration Key | Value Description | Default Value |
|---|---|---|---|
| --nophonehome | disable phonehome in GotCloud and the tools it calls | ||
| BAMUTIL_THINNING | thinning parameter for bamUtil programs (will be set to 0 - if --nophonehome is specified) | --phoneHomeThinning 10 |
Directory Settings Options
These values set GotCloud output subdirectories (relative paths under the OUT_DIR directory). You should not need to change these from the defaults unless you want to use different sub-directory names.
| Configuration Key | Value Description | Default Value |
|---|---|---|
| BAM_GLF_DIR | GLF outputs per BAM (if multiple BAMs per sample) (intermediate files) | glfs/bams |
| SM_GLF_DIR | GLF outputs per sample (intermediate files) | glfs/samples |
| VCF_DIR | unfiltered and filtered VCFs | vcfs |
| PVCF_DIR | vcfPileup results (intermediate files) | pvcfs |
| SPLIT_DIR | VCFs with PASS variants only & split into multiple files | split |
| BEAGLE_DIR | beagle output | beagle |
| SPLIT4_DIR | VCFs with PASS variants only & split into multiple files for running beagle4 | split4 |
| BEAGLE4_DIR | beagle version 4 output | beagle4 |
| THUNDER_DIR | thunder output | thunder |
| TARGET_DIR | directory to store target information when running with a BED file | target |
| GLF_INDEX | filename for index file needed for glfflex (file is created by GotCloud) | glfIndex.ped |
Tool Options
These values set the binaries GotCloud should use. You should not need to change these from the defaults unless you want to try a different version of one of the tools.
Some tools have the options specified with the binary command, while others have them separate or hard coded
| Configuration Key | Program Description | Default Value |
|---|---|---|
| SAMTOOLS_FOR_PILEUP | samtools to use for pileup | $(BIN_DIR)/samtools-hybrid |
| SAMTOOLS_FOR_OTHERS | samtools to use for view and calmd | $(BIN_DIR)/samtools-hybrid |
| GLFMERGE | merge glf files when there are multiple BAMs per indvidual | $(BIN_DIR)/glfMerge |
| GLFFLEX | perform glf-based variant calling (replacement for glfMultiples) | $(BIN_DIR)/glfFlex --minMapQuality 0 --minDepth 1 --maxDepth 10000000 --uniformTsTv --smartFilter |
| VCFPILEUP | vcfPileup to generate rich per-site information | $(BIN_DIR)/vcfPileup |
| INFOCOLLECTOR | gather filtering statistics | $(BIN_DIR)/infoCollector |
| VCFMERGE | merge multiple VCFs separated by chunk of genomes | perl $(SCRIPT_DIR)/bams2vcfMerge.pl |
| VCFCOOKER | vcfCooker program for filtering | $(BIN_DIR)/vcfCooker |
| VCFSUMMARY | script to generate summary statistics of discovered sites | perl $(SCRIPT_DIR)/vcf-summary |
| VCFSPLIT | splits VCF into overlapping chunks for genotype refinement | perl $(SCRIPT_DIR)/vcfSplit.pl |
| VCFSPLIT4 | splits VCF into overlapping chunks for beagle version 4 genotype refinement | perl $(SCRIPT_DIR)/vcfSplit4.pl |
| VCF_SPLIT_CHROM | splits VCF into per chromosome VCFs | perl $(SCRIPT_DIR)/vcfSplitChr.pl |
| VCFPASTE | generate filtered genotype VCF | perl $(SCRIPT_DIR)/vcfPaste.p |
| BEAGLE | beagle program | java -Xmx4g -jar $(BIN_DIR)/beagle.20101226.jar seed=993478 gprobs=true niterations=50 lowmem=true |
| BEAGLE4 | beagle version 4 program | java -Xmx4g -jar $(BIN_DIR)/b4.r1219.jar seed=993478 gprobs=true |
| VCF2BEAGLE | convert VCF (with PL tag) into beagle input | perl $(SCRIPT_DIR)/vcf2Beagle.pl --PL |
| BEAGLE2VCF | convert beagle output to VCF | perl $(SCRIPT_DIR)/beagle2Vcf.pl |
| SVM_SCRIPT | SVM script | perl $(SCRIPT_DIR)/run_libsvm.pl |
| SVMLEARN | SVM program | $(BIN_DIR)/svm-train |
| SVMCLASSIFY | SVM program | $(BIN_DIR)/svm-predict |
| INVNORM | SVM program | $(BIN_DIR)/invNorm |
| THUNDER_STATES | flags for thunder states and weighted states | --states 400 --weightedStates 300 |
| THUNDER | MaCH/Thunder genotype refinement step | $(BIN_DIR)/thunderVCF -r 30 --phase --dosage --compact --inputPhased $(THUNDER_STATES) |
| LIGATEVCF | ligate multiple phased VCFs while resolving the phase between VCFs | perl $(SCRIPT_DIR)/ligateVcf.pl |
| LIGATEVCF4 | ligate multiple phased VCFs while resolving the phase between VCFs | perl $(SCRIPT_DIR)/ligateVcf4.pl |
| VCFCAT | concatenate multiple VCFs | perl $(SCRIPT_DIR)/vcfCat.pl |
| BGZIP | bgzip program | $(BIN_DIR)/bgzip |
| TABIX | tabix program | $(BIN_DIR)/tabix |
| BAMUTIL | bam util program | $(BIN_DIR)/bam |
Options
| Configuration Key | Program Description | Default Value | |
|---|---|---|---|
| SLEEP_MULT | add sleep time prior to some steps; use only if too many steps are starting at the same time doing the same thing | 0 | |
| REMOTE_PREFIX | add a prefix to paths when sending across to a remote machine |
GlfFlex Options
| Configuration Key | Program Description | Default Value |
|---|---|---|
| WGS_SVM | whether or not to run SVM on the whole genome rather than by chromosome (default is by chromosome). Set to TRUE if you are running with a small number of target regions. | |
| VCF_EXTRACT | position file to use for glfFlex | |
| MODEL_GLFSINGLE | set to TRUE if glfSingle model should be used for glfFlex | |
| MODEL_SKIP_DISCOVER | set to true to disable variant discovery for glfFlex | |
| MODEL_AF_PRIOR | set to true to use AF prior for genotyping for glfFlex |
SVM Filtering Options
| Configuration Key | Program Description | Default Value |
|---|---|---|
| POS_SAMPLE | percentage of positive samples used for training | 100 |
| NEG_SAMPLE | percentage of negative samples used for training | 100 |
| SVM_CUTOFF | SVM score cutoff for PASS/FAIL | 0 |
| USE_SVMMODEL | whether to use pre-trained model for SVM filtering | FALSE |
| SVMMODEL | pre-trained model file (if USE_SVMMODEL is set to TRUE) |
Hard Filtering Options
These options set the values to use when applying hard filters.
- To remove any filter, set it to blank in your configuration file
For additional hard filter information, see: GotCloud: Filters
Basic per variant filters:
| Filter | Configuration Key | VCF value checked | Filter Variants with... | Default Value |
|---|---|---|---|---|
| max depth | FILTER_MAX_SAMPLE_DP | INFO:DP | > conf value * total number of samples | 1000[[ |
| min depth | FILTER_MIN_SAMPLE_DP | < conf value * total number of samples | 1 | |
| number of samples with coverage | FILTER_MIN_NS_FRAC | INFO:NS | < conf value * total number of samples | .50 |
| FILTER_MIN_NS | < conf value |
Per variant filters that allow a range of values:
- values of these filters must be numbers (or comma/space separated list of numbers)
- Rules:
- Specifying 1 value in the filter will turn that filter on and use that value
- Specifying 2 values in the filter (separated by ',' and/or ' ') turns on the filter
- Use the 1st value if the number of samples is below FILTER_FORMULA_MIN_SAMPLES
- Use the 2nd value if the number of samples is above FILTER_FORMULA_MAX_SAMPLES
- If the number of samples is between the MIN & MAX, a logscale is used:
(minVal - maxVal) * (log(maxSamples) - log(numSamples)) / (log(maxSamples) - log(minSamples)) + maxVal
| Configuration settings for min/max # samples to determine filter value when the filter setting contains multiple values separated by ',' or ' ' | ||
|---|---|---|
| Configuration Key | Description | Default Value |
| FILTER_FORMULA_MIN_SAMPLES | total number of samples < conf value, use the value before the ',' or ' ' | 100 |
| FILTER_FORMULA_MAX_SAMPLES | total number of samples > conf value, use the value after the ',' or ' ' | 1000 |
| total number of samples between min & max, use logscale | ||
| Filters | |||||
|---|---|---|---|---|---|
| Filter | Configuration Key | VCF value checked | Filter Variants with... | Default Value | Conf Value Requirements |
| max Allele Balance in Heterozygotes | FILTER_MAX_ABL | INFO:AB | > conf value/100.0 | 70,65 | < 100 |
| max Strand Bias Pearson's Correlation | FILTER_MAX_STR | INFO:STR | > conf value/100.0 | 20, 10 | < 100 |
| min Strand Bias Pearson's Correlation | FILTER_MIN_STR | < conf value/100.0 | -20, -10 | > -100 | |
| distance from known indel | FILTER_WIN_INDEL | position | distance from known indel < conf value | 5 | > 0 |
| max Strand Bias z-score | FILTER_MAX_STZ | INFO:STZ | > conf value | 5, 10 | < INT_MAX |
| min Strand Bias z-score | FILTER_MIN_STZ | < conf value | -5, -10 | > INT_MIN | |
| max Alternate allele inflation score | FILTER_MAX_AOI | INFO:AOI | > conf value | 5 | < INT_MAX |
| min FIC | FILTER_MIN_FIC | INFO:FIC | < conf value/100.0 | -20, -10 | > INT_MIN |
| max Cycle Bias Peason's correlation | FILTER_MAX_CBR | INFO:CBR | > conf value/100.0 | 20, 10 | < 100 |
| max LQR | FILTER_MAX_LQR | INFO:LQR | > conf value/100.0 | 30, 20 | < 100 |
| min pred-scaled quality score | FILTER_MIN_QUAL | QUAL | < conf value | 5 | > 0 |
| min Root Mean Squared Mapping Quality | FILTER_MIN_MQ | INFO:MQ | < conf value | 20 | > 0 |
| max Fraction of bases with mapQ=0 | FILTER_MAX_MQ0 | INFO:MQ0 | > conf value/100.0 | 10 | < 100 |
| max Alternate allele quality z-score | FILTER_MAX_AOZ | INFO:AOZ | > conf value | < INT_MAX | |
| max Ratio of base-quality inflation | FILTER_MAX_IOR | INFO:IOR | > conf value | < INT_MAX | |
Additional VCF Cooker filters:
- If you want to add any additional VCF Cooker filters that don't already have a configuration item, you can do that by adding the vcfCooker command-line filter to GotCloud:
| Configuration Key | Default Value |
|---|---|
| FILTER_ADDITIONAL |
Additional Options
| Configuration Key | Program Description | Default Value |
|---|---|---|
| SAMTOOLS_VIEW_FILTER | filter settings for samtools view (default filters by mapping quality and flag) | -q 20 -F 0x0704 |
| NOBAQ_SUBSTRINGS | skip the BAQ step if the BAM filename contains the specified space-separated substrings | SOLID |
| BAM_DEPEND | set to true to rerun the pipeline if the BAM files are newer than previously run steps that use them | FALSE |
| MAKE_OPTS | set to add additional makefile options |
Use Cases & Recommended Settings
Single Sample Processing
To run single sample processing we recommend adding the following settings to your configuration file:
UNIT_CHUNK = 20000000 MODEL_GLFSINGLE = TRUE MODEL_SKIP_DISCOVER = FALSE MODEL_AF_PRIOR = TRUE VCF_EXTRACT = $(REF_DIR)/snpOnly.vcf.gz EXT = $(REF_DIR)/ALL.chrCHR.phase3.combined.sites.unfiltered.vcf.gz $(REF_DIR)/chrCHR.filtered.sites.vcf.gz
Explanation of these settings:
UNIT_CHUNK- since this is only 1 sample, process larger regions at a time than defaultMODEL_GLFSINGLE- single sample, so model glfsingleMODEL_SKIP_DISCOVER- do not skip the variant discovery stepMODEL_AF_PRIOR- use AF prior for genotypingVCF_EXTRACT- VCF file to use for extracting the site information to genotype- This file is included in the latest reference release: hs37d5-db142
EXT- VCF reference files to use for the external filtering- These files are included in the latest reference release: hs37d5-db142
Running
Running variant calling is straightforward:
gotcloud snpcall --conf vc.conf --numjobs 2
gotcloud ldrefine --conf vc.conf --numjobs 2
- Replace
vc.confwith the path/name of the user's configuration file- If you are not overriding any defaults, you can alternatively specify
--list path/bam.listreplacingpath/bam.listwith the path/name of your BAM list file.
- If you are not overriding any defaults, you can alternatively specify
- Replace
2following--numjobswith the number of jobs to be run in parallel - If
OUT_DIRis not defined in the configuration file, add--outdirfollowed by the path to the user's desired output directory.
Running on a Cluster
See Cluster Configuration for information on how to configure GotCloud to run on a cluster.
Results
If there is a failure, you should see a message like:
make: *** [...] Error 1
Where ... is filled in with other text indicating what step failed.
On SNP Call success, you should see the following output sub-directories under your output directory:
- glfs with a bams & samples subdirectory
- pvcfs with a subdirectory per chromosome and then per region
- split with a subdirectory per chromosome
- vcfs with a subdirectory per chromosome
- (optionally your target directory)
Under the vcf/chrXX directory, there should be:
- chrXX.filtered.sites.vcf
- chrXX.filtered.sites.vcf.norm.log
- chrXX.filtered.sites.vcf.summary
- chrXX.filtered.vcf.gz - final filtered variant call file
- chrXX.filtered.vcf.gz.OK
- chrXX.filtered.vcf.gz.tbi
- chrXX.hardfiltered.sites.vcf
- chrXX.hardfiltered.sites.vcf.log
- chrXX.hardfiltered.sites.vcf.summary
- chrXX.hardfiltered.vcf.gz
- chrXX.hardfiltered.vcf.gz.OK
- chrXX.hardfiltered.vcf.gz.tbi
- chrXX.merged.sites.vcf
- chrXX.merged.stats.vcf
- chrXX.merged.vcf
- chrXX.merged.vcf.OK
The .merged.vcf is the merged together versions of the separate regions in the same chromosome.
The filtered is the merged.vcf after it has been run through filters and is marked with PASS/FAIL.
Under the split/chrXX directory, there should be:
- chrXX.filtered.PASS.split.[N].vcf.gz
- chrXX.filtered.PASS.split.err
- chrXX.filtered.PASS.split.vcflist
- chrXX.filtered.PASS.gz - final variant call file with only PASS variants
- subset.OK