LASER
Introduction
LASER, which stands for Locating Ancestry using SEquencing Reads, is a C++ software package that can estimate individual ancestry directly from genome-wide shortgun sequencing reads without calling genotypes. The method relies on the availability of a set of reference individuals whose genome-wide SNP genotypes and ancestral information are known. We first construct a reference coordinate system by applying principal components analysis (PCA) to the genotype data of the reference individuals. Then, for each sequencing sample, use the genome-wide sequencing reads to place the sample into the reference PCA space. With an appropriate reference panel, the estimated coordinates of the sequencing samples identify their ancestral background and can be directly used to correct for population structure in association studies or to ensure adequate matching of cases and controls.
Note:
The goal of this wiki page is to help you get start using LASER.
This page was created for LASER 1.0. Some of the information might be outdated for LASER 2.0.
A more updated wiki page can be found at 2014 UM Sequencing Workshop.
We also encourage you to read the manual for more details of the software.
Download
To get a copy of the software and manual, go to the LASER Download page.
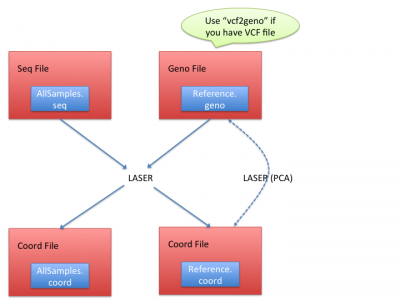
Workflow
LASER generates the coordinates from both reference individuals and sequence samples. It requires essentially two input files:
- Seq file: a text file processed from BAM (alignment) files. (See Processing sequencing file for how to prepare seq file)
- Geno file: genotypes of reference individuals. (See Geno file to understand geno file format)
LASER typically outputs two coord files: (1) in reference individuals' coord file(Reference.coord), LASER outputs the reference coordinates in the PCA space; (2) in sequence samples' coord files(AllSamples.coord), LASER infers their ancestries by placing their ancestry coordinates onto reference samples' PCA space.
An example result of the coord file of sequence samples is shown below:
popID indivID L1 Ci t PC1 PC2 YRI NA19238 1409 0.304122 0.98933 52.7634 -39.7924 CEU NA12892 1552 0.330037 0.989709 9.82674 25.2898 CEU NA12891 1609 0.362198 0.988082 0.439573 26.8872 CEU NA12878 1579 0.334825 0.988677 8.83775 28.1342 YRI NA19239 1558 0.34898 0.988302 53.9104 -39.1727 YRI NA19240 1735 0.404142 0.990264 59.8379 -45.2765
In the header line, popID means "population ID", indivID means "individual ID", L1 means number of loci that has been covered by at least one read, Ci means "average coverage", t means Procrustes similarity. PC1 and PC2 mean coordinates of the first and second principal components.
Tutorial
In this tutorial, we will show you how to prepare data and run LASER.
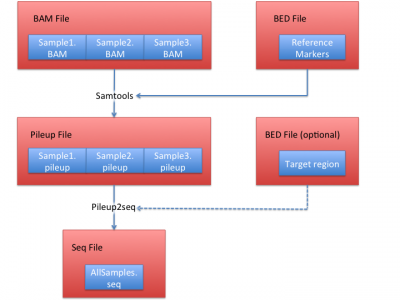
Process sequencing file (BAM)
We illustrate how to obtain .seq file from BAM files in this section. In this example, we use HGDP data set as a reference, which contains 938 individuals and 632,958 markers.
1. Obtain pileup files from BAM files
The first step is to generate a BED file:
cat ../resource/HGDP/HGDP_938.site |awk '{if (NR > 1) {print $1, $2-1, $2;}}' > HGDP_938.bed
This BED file contains the positions of all the reference markers.
Then we use samtools to extract the sequence bases overlapping these 632,958 reference markers. Assuming your BAM file name is NA12878.chrom22.recal.bam (our example BAM file), you can use this:
samtools mpileup -q 30 -Q 20 -f ../../LASER-resource/reference/hs37d5.fa -l HGDP_938.bed exampleBAM/NA12878.chrom22.recal.bam > NA12878.chrom22.pileup
to obtain a pileup file named NA12878.chrom22.pileup. It is required to keep the .pileup' suffix.
2. Obtain a seq file from pileup files.
After obtaining pileup files from each BAM file, you can convert them into a single seq file before running LASER. Use the same site file and all generated pileup files from step 1 to generate a seq file:
python pileup2seq.py -m ../resource/HGDP/HGDP_938.site -o test NA12878.chrom22.pileup
You should obtain test.seq file after this step.
Estimate ancestries of sequence samples
The easiest way to perform LASER using its exemplar data is:
./laser -s pileup2seq/test.seq -g resource/HGDP/HGDP_938.geno -c resource/HGDP/HGDP_938.RefPC.coord -o test -k 2
Upon successful calculation, you will find a result file "test.SeqPC.coord".
Interpret LASER outputs
Upon successfully launching LASER command line as above, the output messages should be similar to below:
=================================================================== ==== LASER: Locating Ancestry from SEquencing Reads ==== ==== Version 1.0 | (c) Chaolong Wang 2013 ==== ==================================================================== Started at: Fri Nov 15 01:05:48 2013
938 individuals are detected in the GENO_FILE. 632958 loci are detected in the GENO_FILE. 1 individuals are detected in the SEQ_FILE. 632958 loci are detected in the SEQ_FILE. 938 individuals are detected in the COORD_FILE. 100 PCs are detected in the COORD_FILE.
Parameter values used in execution: ------------------------------------------------- GENO_FILE (-g)resource/HGDP/HGDP_938.geno SEQ_FILE (-s)pileup2seq/test.seq COORD_FILE (-c)resource/HGDP/HGDP_938.RefPC.coord OUT_PREFIX (-o)test DIM (-k)2 MIN_LOCI (-l)100 SEQ_ERR (-e)0.01 FIRST_IND (-x)1 LAST_IND (-y)1 REPS (-r)1 OUTPUT_REPS (-R)0 CHECK_FORMAT (-fmt)10 CHECK_COVERAGE (-cov)0 PCA_MODE (-pca)0 -------------------------------------------------
Fri Nov 15 01:05:50 2013 Checking data format ... GENO_FILE: OK. SEQ_FILE: OK. COORD_FILE: OK.
Fri Nov 15 01:06:01 2013 Reading reference genotypes ...
Fri Nov 15 01:09:15 2013 Reading reference PCA coordinates ...
Fri Nov 15 01:09:15 2013 Analyzing sequence samples ... Results for the sequence samples are output to 'test.SeqPC.coord'.
Finished at: Fri Nov 15 01:09:21 2013 ====================================================================
The ancestry of input samples are store in the file test.SeqPC.coord, which content is shown below:
popID indivID L1 Ci t PC1 PC2 NA12878.chrom22 NA12878.chrom22 1601 0.00858193 0.977243 31.522 224.098
The ancestry coordinates for NA12878 samples are given in PC1 (31.522) and PC2 (224.098).
It is recommended to visualize this results with HGDP reference samples whose coordinates are given in file: resource/HGDP/HGDP_938.RefPC.coord
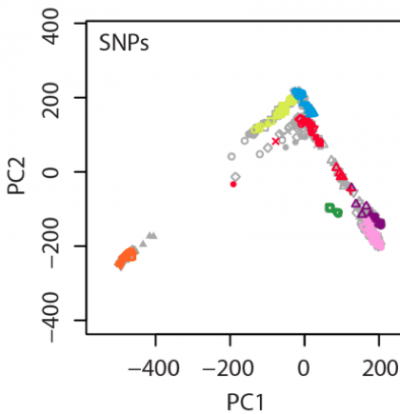
In our manuscript, an example figure is shown:
In this figure, 238 individuals were randomly selected from the total 938 HGDP samples as the testing set (colored symbols), and the remaining 700 HGDP individuals were used as the reference panel (gray symbols).
File format
Geno file
Geno file are from reference samples. LASER use genotype of these samples as a reference panel. You can obtain geno file from VCF files using vcf2geno.
In our resource folder, we provide an example geno file for the HGDP data set (resource/HGDP/HGDP_938.geno):
Brahui HGDP00001 1 2 1 1 0 2 0 2 1 2 2 2 1 1 2 1 0 Brahui HGDP00003 0 0 2 0 0 2 0 2 0 2 2 2 2 0 2 2 0 Brahui HGDP00005 0 2 2 0 0 1 0 2 1 2 2 2 2 1 2 2 1 Brahui HGDP00007 0 2 2 0 0 2 0 2 0 2 2 2 1 1 2 2 1 Brahui HGDP00009 0 1 0 1 0 2 0 2 0 2 2 2 2 0 2 2 0 Brahui HGDP00011 1 1 2 1 1 2 1 1 1 2 2 2 1 1 2 2 0 Brahui HGDP00013 1 2 2 1 1 2 1 2 0 2 2 2 2 0 2 2 0 Brahui HGDP00015 1 1 2 0 0 2 0 2 0 2 2 2 2 0 2 2 0 Brahui HGDP00017 1 1 2 0 0 1 0 0 0 2 0 1 1 2 2 2 0 Brahui HGDP00019 0 2 2 0 0 1 0 1 0 2 1 2 2 1 2 2 0
The first and second columns represent the population id and individual id. From the third column, each number represents a genotype. To be consistent with the sequence data, genotypes should be given on the forward strand. Genotypes are coded by 0, 1, or 2, representing copies of the reference allele at a locus in one individual.
In this geno file, we have 632,960 columns which contains 632,958 markers from column 3 to the last column.
Seq file
Seq file is generated from pileup files. It contains sequencing information and organize it in a LASER readable format. The first two columns represent population id and individual id. Subsequent columns are total read depths and reference base counts. For example, column 3 and 4 are 0, 0 in the following example. That means at first marker, the sequence read depth is 0 and thus none of the reads has reference base. We enforce tab delimiters between markers and space delimiters between each read depths and reference base counts. On line of seq file looks like below:
NA12878.chrom22 NA12878.chrom22 0 0 0 0 0 0 0 0 0
Pileup file
Pileup file are generated using samtools. An example pileup file is shown below:
22 17094749 A 1 c D 22 17202602 T 1 . D 22 17411899 A 1 . C 22 17450515 G 2 ., 9< 22 17452966 T 1 c 5 22 17470779 C 1 , A 22 17492203 G 1 , B 22 17504945 C 3 ,.. BCA 22 17529814 T 3 .., CCC
The columns are chromosome, position (1-based), reference base, depth, bases and base qualities.
BED file
BED file represents genomic regions and it follows UCSC conventions:
1 752565 752566 1 768447 768448 1 1005805 1005806 1 1018703 1018704 1 1021414 1021415
The columns are: chromosome, start position (0-based) and end position (1-based).
Coord file
Coord files represent the ancestries of both reference samples and sequence samples. An example coord file looks like below:
popID indivID L1 Ci t PC1 PC2 YRI NA19238 1409 0.304122 0.98933 52.7634 -39.7924 CEU NA12892 1552 0.330037 0.989709 9.82674 25.2898 CEU NA12891 1609 0.362198 0.988082 0.439573 26.8872 CEU NA12878 1579 0.334825 0.988677 8.83775 28.1342 YRI NA19239 1558 0.34898 0.988302 53.9104 -39.1727 YRI NA19240 1735 0.404142 0.990264 59.8379 -45.2765
The columns are: popID means "population ID", indivID means "individual ID", L1 means number of loci has been covered, Ci means "average coverage", t means Procrustes similarity. PC1, PC2 means coordinates of first and second principal components. You may notice L1, Ci, and t are omitted in the coord files of reference samples. The reason is that reference samples use genotypes and do not have coverage information.
Site file
Site file is equivalent to BED file and it is used here to represent marker positions. An example site file looks like below:
CHR POS ID REF ALT 1 752566 rs3094315 G A 1 768448 rs12562034 G A 1 1005806 rs3934834 C T 1 1018704 rs9442372 A G 1 1021415 rs3737728 A G
The site file has header line, and it contains chromosome, position(1-based), id (usually marker name), ref (reference allele) and alt (alternative allele).
Advanced options
LASER has advanced options including (1) parallel computing; (2) increase ancestry inference accuracy using repeated runs; (3) generate PCA coordiates using genotypes. See LASER Manual for detailed information.
Contact
Comments on this wiki page or questions related to preparing input files for LASER can be sent to Xiaowei Zhan. Comments on the LASER software or the user's manual can be sent to Chaolong Wang. This project was directed by Gonçalo Abecasis and Sebastian Zöllner at the University of Michigan.